Human CalDAG-GEFI gene (RASGRP2) mutation affects platelet function and causes severe bleeding
- PMID: 24958846
- PMCID: PMC4076591
- DOI: 10.1084/jem.20130477
Human CalDAG-GEFI gene (RASGRP2) mutation affects platelet function and causes severe bleeding
Abstract
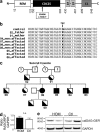
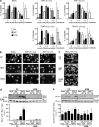
The nature of an inherited platelet disorder was investigated in three siblings affected by severe bleeding. Using whole-exome sequencing, we identified the culprit mutation (cG742T) in the RAS guanyl-releasing protein-2 (RASGRP2) gene coding for calcium- and DAG-regulated guanine exchange factor-1 (CalDAG-GEFI). Platelets from individuals carrying the mutation present a reduced ability to activate Rap1 and to perform proper αIIbβ3 integrin inside-out signaling. Expression of CalDAG-GEFI mutant in HEK293T cells abolished Rap1 activation upon stimulation. Nevertheless, the PKC- and ADP-dependent pathways allow residual platelet activation in the absence of functional CalDAG-GEFI. The mutation impairs the platelet's ability to form thrombi under flow and spread normally as a consequence of reduced Rac1 GTP-binding. Functional deficiencies were confined to platelets and megakaryocytes with no leukocyte alteration. This contrasts with the phenotype seen in type III leukocyte adhesion deficiency caused by the absence of kindlin-3. Heterozygous did not suffer from bleeding and have normal platelet aggregation; however, their platelets mimicked homozygous ones by failing to undergo normal adhesion under flow and spreading. Rescue experiments on cultured patient megakaryocytes corrected the functional deficiency after transfection with wild-type RASGRP2. Remarkably, the presence of a single normal allele is sufficient to prevent bleeding, making CalDAG-GEFI a novel and potentially safe therapeutic target to prevent thrombosis.
© 2014 Canault et al.
Figures





Comment in
-
Identification of a severe bleeding disorder in humans caused by a mutation in CalDAG-GEFI.J Exp Med. 2014 Jun 30;211(7):1271. doi: 10.1084/jem.2117insight1. J Exp Med. 2014. PMID: 24980744 Free PMC article. No abstract available.
References
-
- Alon R., Aker M., Feigelson S., Sokolovsky-Eisenberg M., Staunton D.E., Cinamon G., Grabovsky V., Shamri R., Etzioni A. 2003. A novel genetic leukocyte adhesion deficiency in subsecond triggering of integrin avidity by endothelial chemokines results in impaired leukocyte arrest on vascular endothelium under shear flow. Blood. 101:4437–4445 10.1182/blood-2002-11-3427 - DOI - PubMed
-
- Bergmeier W., Goerge T., Wang H.W., Crittenden J.R., Baldwin A.C., Cifuni S.M., Housman D.E., Graybiel A.M., Wagner D.D. 2007. Mice lacking the signaling molecule CalDAG-GEFI represent a model for leukocyte adhesion deficiency type III. J. Clin. Invest. 117:1699–1707 10.1172/JCI30575 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

