Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi
- PMID: 24958869
- PMCID: PMC4103376
- DOI: 10.1073/pnas.1400592111
Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi
Erratum in
- Proc Natl Acad Sci U S A. 2014 Oct 14;111(41):14959
Abstract
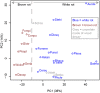
Basidiomycota (basidiomycetes) make up 32% of the described fungi and include most wood-decaying species, as well as pathogens and mutualistic symbionts. Wood-decaying basidiomycetes have typically been classified as either white rot or brown rot, based on the ability (in white rot only) to degrade lignin along with cellulose and hemicellulose. Prior genomic comparisons suggested that the two decay modes can be distinguished based on the presence or absence of ligninolytic class II peroxidases (PODs), as well as the abundance of enzymes acting directly on crystalline cellulose (reduced in brown rot). To assess the generality of the white-rot/brown-rot classification paradigm, we compared the genomes of 33 basidiomycetes, including four newly sequenced wood decayers, and performed phylogenetically informed principal-components analysis (PCA) of a broad range of gene families encoding plant biomass-degrading enzymes. The newly sequenced Botryobasidium botryosum and Jaapia argillacea genomes lack PODs but possess diverse enzymes acting on crystalline cellulose, and they group close to the model white-rot species Phanerochaete chrysosporium in the PCA. Furthermore, laboratory assays showed that both B. botryosum and J. argillacea can degrade all polymeric components of woody plant cell walls, a characteristic of white rot. We also found expansions in reducing polyketide synthase genes specific to the brown-rot fungi. Our results suggest a continuum rather than a dichotomy between the white-rot and brown-rot modes of wood decay. A more nuanced categorization of rot types is needed, based on an improved understanding of the genomics and biochemistry of wood decay.
Keywords: bioenergy; lignocellulose; phylogenomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Kirk PMCP, Minter DW, Stalpers JA. Dictionary of the Fungi. 10th Ed. Wallingford, UK: CAB International; 2008. p. 72.
-
- Johansson M, Stenlid J. Infection of roots of Norway spruce (Picea abies) by Heterobasidion Annosum: Initial reactions in sapwood by wounding and infection. Eur J Forest Pathol. 1985;15(1):32–45.
-
- Otrosina WJ, Chase TE, Cobb FW, Korhonen K. Population structure of Heterobasidion annosum from North America and Europe. Can J Bot. 1993;71(8):1064–1071.
-
- Martin F, et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452(7183):88–92. - PubMed
-
- Jin Y, et al. Characterization of seedling infection types and adult plant infection responses of monogenic Sr gene lines to race TTKS of Puccinia graminis f. sp tritici. Plant Dis. 2007;91(9):1096–1099. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

