Leishmania spp. Proteome Data Sets: A Comprehensive Resource for Vaccine Development to Target Visceral Leishmaniasis
- PMID: 24959165
- PMCID: PMC4050426
- DOI: 10.3389/fimmu.2014.00260
Leishmania spp. Proteome Data Sets: A Comprehensive Resource for Vaccine Development to Target Visceral Leishmaniasis
Abstract
Visceral leishmaniasis is a neglected infectious disease caused primarily by Leishmania donovani and Leishmania infantum protozoan parasites. A significant number of infections take a fatal course. Drug therapy is available but still costly and parasites resistant to first line drugs are observed. Despite many years of trial no commercial vaccine is available to date. However, development of a cost effective, needle-independent vaccine remains a high priority. Reverse vaccinology has attracted much attention since the term has been coined and the approach tested by Rappuoli and colleagues. This in silico selection of antigens from genomic and proteomic data sets was also adapted to aim at developing an anti-Leishmania vaccine. Here, an analysis of the efforts is attempted and the challenges to be overcome by these endeavors are discussed. Strategies that led to successful identification of antigens will be illustrated. Furthermore, these efforts are viewed in the context of anticipated modes of action of effective anti-Leishmania immune responses to highlight possible advantages and shortcomings.
Keywords: T cell antigen receptor; kinetoplastida; major histocompatibility complex antigens; proteome; vaccine.
Figures
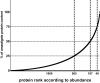
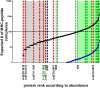
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

