Kelch proteins: emerging roles in skeletal muscle development and diseases
- PMID: 24959344
- PMCID: PMC4067060
- DOI: 10.1186/2044-5040-4-11
Kelch proteins: emerging roles in skeletal muscle development and diseases
Abstract
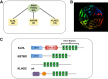
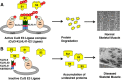
Our understanding of genes that cause skeletal muscle disease has increased tremendously over the past three decades. Advances in approaches to genetics and genomics have aided in the identification of new pathogenic mechanisms in rare genetic disorders and have opened up new avenues for therapeutic interventions by identification of new molecular pathways in muscle disease. Recent studies have identified mutations of several Kelch proteins in skeletal muscle disorders. The Kelch superfamily is one of the largest evolutionary conserved gene families. The 66 known family members all possess a Kelch-repeat containing domain and are implicated in diverse biological functions. In skeletal muscle development, several Kelch family members regulate the processes of proliferation and/or differentiation resulting in normal functioning of mature muscles. Importantly, many Kelch proteins function as substrate-specific adaptors for Cullin E3 ubiquitin ligase (Cul3), a core component of the ubiquitin-proteasome system to regulate the protein turnover. This review discusses the emerging roles of Kelch proteins in skeletal muscle function and disease.
Keywords: BACK; BTB; Congenital myopathy; Cul3; Differentiation; Dystrophy; Kelch; Nemaline myopathy; Proliferation; Proteasome; Skeletal muscle; Ubiquitination.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

