HomeRun Vector Assembly System: a flexible and standardized cloning system for assembly of multi-modular DNA constructs
- PMID: 24959875
- PMCID: PMC4069157
- DOI: 10.1371/journal.pone.0100948
HomeRun Vector Assembly System: a flexible and standardized cloning system for assembly of multi-modular DNA constructs
Abstract
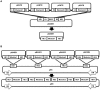
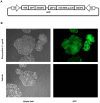
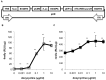
Advances in molecular and synthetic biology call for efficient assembly of multi-modular DNA constructs. We hereby present a novel modular cloning method that obviates the need for restriction endonucleases and significantly improves the efficiency in the design and construction of complex DNA molecules by standardizing all DNA elements and cloning reactions. Our system, named HomeRun Vector Assembly System (HVAS), employs a three-tiered vector series that utilizes both multisite gateway cloning and homing endonucleases, with the former building individual functional modules and the latter linking modules into the final construct. As a proof-of-principle, we first built a two-module construct that supported doxycycline-induced expression of green fluorescent protein (GFP). Further, with a three-module construct we showed quantitatively that there was minimal promoter leakage between neighbouring modules. Finally, we developed a method, in vitro Cre recombinase-mediated cassette exchange (RMCE) cloning, to regenerate a gateway destination vector from a previous multisite gateway cloning reaction, allowing access to existing DNA element libraries in conventional gateway entry clones, and simple creation of constructs ready for in vivo RMCE. We believe these methods constitute a useful addition to the standard molecular cloning techniques that could potentially support industrial scale synthesis of DNA constructs.
Conflict of interest statement
Figures

References
-
- Knight T (2003) Idempotent vector design for standard assembly of biobricks. MIT Synthetic Biology Working Group Technical Reports, http://hdl.handle.net/1721.1/21168.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

