RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing
- PMID: 24960161
- PMCID: PMC4109538
- DOI: 10.1172/JCI74523
RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing
Abstract
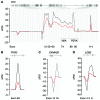
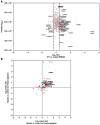
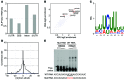
Mutations in the gene encoding the RNA-binding protein RBM20 have been implicated in dilated cardiomyopathy (DCM), a major cause of chronic heart failure, presumably through altering cardiac RNA splicing. Here, we combined transcriptome-wide crosslinking immunoprecipitation (CLIP-seq), RNA-seq, and quantitative proteomics in cell culture and rat and human hearts to examine how RBM20 regulates alternative splicing in the heart. Our analyses revealed the presence of a distinct RBM20 RNA-recognition element that is predominantly found within intronic binding sites and linked to repression of exon splicing with RBM20 binding near 3' and 5' splice sites. Proteomic analysis determined that RBM20 interacts with both U1 and U2 small nuclear ribonucleic particles (snRNPs) and suggested that RBM20-dependent splicing repression occurs through spliceosome stalling at complex A. Direct RBM20 targets included several genes previously shown to be involved in DCM as well as genes not typically associated with this disease. In failing human hearts, reduced expression of RBM20 affected alternative splicing of several direct targets, indicating that differences in RBM20 expression may affect cardiac function. Together, these findings identify RBM20-regulated targets and provide insight into the pathogenesis of human heart failure.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

