KLHL40 deficiency destabilizes thin filament proteins and promotes nemaline myopathy
- PMID: 24960163
- PMCID: PMC4109545
- DOI: 10.1172/JCI74994
KLHL40 deficiency destabilizes thin filament proteins and promotes nemaline myopathy
Abstract
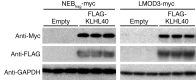
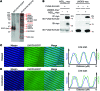
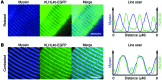
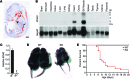
Nemaline myopathy (NM) is a congenital myopathy that can result in lethal muscle dysfunction and is thought to be a disease of the sarcomere thin filament. Recently, several proteins of unknown function have been implicated in NM, but the mechanistic basis of their contribution to disease remains unresolved. Here, we demonstrated that loss of a muscle-specific protein, kelch-like family member 40 (KLHL40), results in a nemaline-like myopathy in mice that closely phenocopies muscle abnormalities observed in KLHL40-deficient patients. We determined that KLHL40 localizes to the sarcomere I band and A band and binds to nebulin (NEB), a protein frequently implicated in NM, as well as a putative thin filament protein, leiomodin 3 (LMOD3). KLHL40 belongs to the BTB-BACK-kelch (BBK) family of proteins, some of which have been shown to promote degradation of their substrates. In contrast, we found that KLHL40 promotes stability of NEB and LMOD3 and blocks LMOD3 ubiquitination. Accordingly, NEB and LMOD3 were reduced in skeletal muscle of both Klhl40-/- mice and KLHL40-deficient patients. Loss of sarcomere thin filament proteins is a frequent cause of NM; therefore, our data that KLHL40 stabilizes NEB and LMOD3 provide a potential basis for the development of NM in KLHL40-deficient patients.
Figures


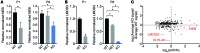

References
-
- Laing NG, et al. A mutation in the α tropomyosin gene TPM3 associated with autosomal dominant nemaline myopathy NEM1. Nat Genet. 1995;10(2):249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL077439/HL/NHLBI NIH HHS/United States
- T32 GM008014/GM/NIGMS NIH HHS/United States
- U01 HL100401/HL/NHLBI NIH HHS/United States
- U01-HL-100401/HL/NHLBI NIH HHS/United States
- R01 HL093039/HL/NHLBI NIH HHS/United States
- R01 DK099653/DK/NIDDK NIH HHS/United States
- DK-099653/DK/NIDDK NIH HHS/United States
- R01 HL111665/HL/NHLBI NIH HHS/United States
- HL-077439/HL/NHLBI NIH HHS/United States
- HL-111665/HL/NHLBI NIH HHS/United States
- HL-093039/HL/NHLBI NIH HHS/United States
- T32 HL007360/HL/NHLBI NIH HHS/United States
- T32-HL-007360/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

