BioC implementations in Go, Perl, Python and Ruby
- PMID: 24961236
- PMCID: PMC4067548
- DOI: 10.1093/database/bau059
BioC implementations in Go, Perl, Python and Ruby
Abstract
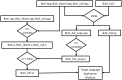
As part of a communitywide effort for evaluating text mining and information extraction systems applied to the biomedical domain, BioC is focused on the goal of interoperability, currently a major barrier to wide-scale adoption of text mining tools. BioC is a simple XML format, specified by DTD, for exchanging data for biomedical natural language processing. With initial implementations in C++ and Java, BioC provides libraries of code for reading and writing BioC text documents and annotations. We extend BioC to Perl, Python, Go and Ruby. We used SWIG to extend the C++ implementation for Perl and one Python implementation. A second Python implementation and the Ruby implementation use native data structures and libraries. BioC is also implemented in the Google language Go. BioC modules are functional in all of these languages, which can facilitate text mining tasks. BioC implementations are freely available through the BioC site: http://bioc.sourceforge.net. Database URL: http://bioc.sourceforge.net/
Published by Oxford University Press 2014. This work is written by US Government employees and is in the public domain in the US.
Figures







References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

