Genome sequence and phenotypic analysis of a first German Francisella sp. isolate (W12-1067) not belonging to the species Francisella tularensis
- PMID: 24961323
- PMCID: PMC4230796
- DOI: 10.1186/1471-2180-14-169
Genome sequence and phenotypic analysis of a first German Francisella sp. isolate (W12-1067) not belonging to the species Francisella tularensis
Abstract
Background: Francisella isolates from patients suffering from tularemia in Germany are generally strains of the species F. tularensis subsp. holarctica. To our knowledge, no other Francisella species are known for Germany. Recently, a new Francisella species could be isolated from a water reservoir of a cooling tower in Germany.
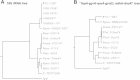
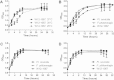
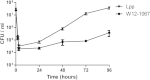
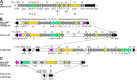
Results: We identified a Francisella sp. (isolate W12-1067) whose 16S rDNA is 99% identical to the respective nucleotide sequence of the recently published strain F. guangzhouensis. The overall sequence identity of the fopA, gyrA, rpoA, groEL, sdhA and dnaK genes is only 89%, indicating that strain W12-1067 is not identical to F. guangzhouensis. W12-1067 was isolated from a water reservoir of a cooling tower of a hospital in Germany. The growth optimum of the isolate is approximately 30°C, it can grow in the presence of 4-5% NaCl (halotolerant) and is able to grow without additional cysteine within the medium. The strain was able to replicate within a mouse-derived macrophage-like cell line. The whole genome of the strain was sequenced (~1.7 mbp, 32.2% G + C content) and the draft genome was annotated. Various virulence genes common to the genus Francisella are present, but the Francisella pathogenicity island (FPI) is missing. However, another putative type-VI secretion system is present within the genome of strain W12-1067.
Conclusions: Isolate W12-1067 is closely related to the recently described F. guangzhouensis species and it replicates within eukaryotic host cells. Since W12-1067 exhibits a putative new type-VI secretion system and F. tularensis subsp. holarctica was found not to be the sole species in Germany, the new isolate is an interesting species to be analyzed in more detail. Further research is needed to investigate the epidemiology, ecology and pathogenicity of Francisella species present in Germany.
Figures

References
-
- Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Fine AD, Friedlander AM, Hauer J, Layton M, Lillibridge SR, McDade JE, Osterholm MT, O'Toole T, Parker G, Perl TM, Russell PK, Tonat K. Working Group on Civilian Biodefense. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285:2763–2773. - PubMed
-
- Wenger JD, Hollis DG, Weaver RE, Baker CN, Brown GR, Brenner DJ, Broome CV. Infection caused by Francisella philomiragia (formerly Yersinia philomiragia). A newly recognized human pathogen. Ann Intern Med. 1989;110:888–892. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

