High-throughput transcriptome sequencing identifies candidate genetic modifiers of vulnerability to fetal alcohol spectrum disorders
- PMID: 24962712
- PMCID: PMC4149215
- DOI: 10.1111/acer.12457
High-throughput transcriptome sequencing identifies candidate genetic modifiers of vulnerability to fetal alcohol spectrum disorders
Abstract
Background: Fetal alcohol spectrum disorders (FASD) is a leading cause of neurodevelopmental disability. Genetic factors can modify vulnerability to FASD, but these elements are poorly characterized.
Methods: We performed high-throughput transcriptional profiling to identify gene candidates that could potentially modify vulnerability to ethanol's (EtOH's) neurotoxicity. We interrogated a unique genetic resource, neuroprogenitor cells from 2 closely related Gallus gallus lines having well-characterized robust or attenuated EtOH responses with respect to intracellular calcium mobilization and CaMKII/β-catenin-dependent apoptosis. Samples were not exposed to EtOH prior to analysis.
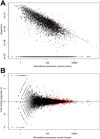
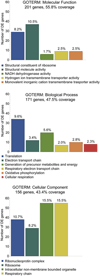
Results: We identified 363 differentially expressed genes in neuroprogenitors from these 2 lines. Kyoto Encyclopedia of Genes and Genomes analysis revealed several gene clusters having significantly differential enrichment in gene expression. The largest and most significant cluster comprised ribosomal proteins (38 genes, p = 1.85 × 10(-47) ). Other significantly enriched gene clusters included metabolism (25 genes, p = 0.0098), oxidative phosphorylation (18 genes, p = 1.10 × 10(-11) ), spliceosome (13 genes, p = 7.02 × 10(-8) ), and protein processing in the endoplasmic reticulum (9 genes, p = 0.0011). Inspection of gene ontogeny (GO) terms identified 24 genes involved in the calcium/β-catenin signals that mediate EtOH's neurotoxicity in this model, including β-catenin itself and both calmodulin isoforms.
Conclusions: Four of the identified pathways with altered transcript abundance mediate the flow of cellular information from RNA to protein. Importantly, ribosome biogenesis also senses nucleolar stress and regulates p53-mediated apoptosis in neural crest. Human ribosomopathies produce craniofacial malformations and 11 known ribosomopathy genes were differentially expressed in this model of neural crest apoptosis. Rapid changes in ribosome expression are consistently observed in EtOH-treated mouse embryo neural folds, a model that is developmentally similar to ours. The recurring identification of ribosome biogenesis suggests it is a candidate modifier of EtOH vulnerability. These results highlight this approach's efficacy to formulate new, mechanistic hypotheses regarding EtOH's developmental damage.
Keywords: Apoptosis; Diamond-Blackfan Anemia; Fetal Alcohol Spectrum Disorders; Gallus gallus; Neural Crest; Oxidative Phosphorylation; Ribosome Biogenesis; Transcriptome Profiling.
Copyright © 2014 by the Research Society on Alcoholism.
Figures

Similar articles
-
Transcriptome Profiling Identifies Ribosome Biogenesis as a Target of Alcohol Teratogenicity and Vulnerability during Early Embryogenesis.PLoS One. 2017 Jan 3;12(1):e0169351. doi: 10.1371/journal.pone.0169351. eCollection 2017. PLoS One. 2017. PMID: 28046103 Free PMC article.
-
An evolutionarily conserved mechanism of calcium-dependent neurotoxicity in a zebrafish model of fetal alcohol spectrum disorders.Alcohol Clin Exp Res. 2014 May;38(5):1255-65. doi: 10.1111/acer.12360. Epub 2014 Feb 11. Alcohol Clin Exp Res. 2014. PMID: 24512079 Free PMC article.
-
Calcium-mediated repression of β-catenin and its transcriptional signaling mediates neural crest cell death in an avian model of fetal alcohol syndrome.Birth Defects Res A Clin Mol Teratol. 2011 Jul;91(7):591-602. doi: 10.1002/bdra.20833. Epub 2011 May 31. Birth Defects Res A Clin Mol Teratol. 2011. PMID: 21630427 Free PMC article.
-
Neural crest development in fetal alcohol syndrome.Birth Defects Res C Embryo Today. 2014 Sep;102(3):210-20. doi: 10.1002/bdrc.21078. Epub 2014 Sep 15. Birth Defects Res C Embryo Today. 2014. PMID: 25219761 Free PMC article. Review.
-
Genomic factors that shape craniofacial outcome and neural crest vulnerability in FASD.Front Genet. 2014 Aug 7;5:224. doi: 10.3389/fgene.2014.00224. eCollection 2014. Front Genet. 2014. PMID: 25147554 Free PMC article. Review.
Cited by
-
Exon level machine learning analyses elucidate novel candidate miRNA targets in an avian model of fetal alcohol spectrum disorder.PLoS Comput Biol. 2019 Apr 11;15(4):e1006937. doi: 10.1371/journal.pcbi.1006937. eCollection 2019 Apr. PLoS Comput Biol. 2019. PMID: 30973878 Free PMC article.
-
New Animal Models for Understanding FMRP Functions and FXS Pathology.Cells. 2022 May 12;11(10):1628. doi: 10.3390/cells11101628. Cells. 2022. PMID: 35626665 Free PMC article. Review.
-
Gene-alcohol interactions in birth defects.Curr Top Dev Biol. 2023;152:77-113. doi: 10.1016/bs.ctdb.2022.10.003. Epub 2022 Nov 14. Curr Top Dev Biol. 2023. PMID: 36707215 Free PMC article.
-
Ribosomal biogenesis as an emerging target of neurodevelopmental pathologies.J Neurochem. 2019 Feb;148(3):325-347. doi: 10.1111/jnc.14576. Epub 2018 Nov 12. J Neurochem. 2019. PMID: 30144322 Free PMC article. Review.
-
Neural crest cells and fetal alcohol spectrum disorders: Mechanisms and potential targets for prevention.Pharmacol Res. 2023 Aug;194:106855. doi: 10.1016/j.phrs.2023.106855. Epub 2023 Jul 17. Pharmacol Res. 2023. PMID: 37460002 Free PMC article. Review.
References
-
- Boehm SL, 2nd, Lundahl KR, Caldwell J, Gilliam DM. Ethanol teratogenesis in the C57BL/6J, DBA/2J, and A/J inbred mouse strains. Alcohol. 1997;14:389–395. - PubMed
-
- Bugner V1, Tecza A, Gessert S, Kühl M. Peter Pan functions independently of its role in ribosome biogenesis during early eye and craniofacial cartilage development in Xenopus laevis. Development. 2011;138:2369–2378. - PubMed
-
- Carter RC, Jacobson SW, Molteno CD, Jacobson JL. Fetal alcohol exposure, iron-deficiency anemia, and infant growth. Pediatrics. 2007;120:559–567. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

