Comparative Analysis of Miscanthus and Saccharum Reveals a Shared Whole-Genome Duplication but Different Evolutionary Fates
- PMID: 24963058
- PMCID: PMC4114942
- DOI: 10.1105/tpc.114.125583
Comparative Analysis of Miscanthus and Saccharum Reveals a Shared Whole-Genome Duplication but Different Evolutionary Fates
Abstract
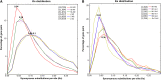
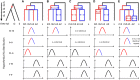
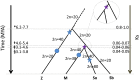
Multiple polyploidizations with divergent consequences in the grass subtribe Saccharinae provide a singular opportunity to study in situ adaptation of a genome to the duplicated state, heretofore known primarily from paleogenomics. We show that allopolyploidy in a common Miscanthus-Saccharum ancestor ∼3.8 to 4.6 million years ago closely coincides in time with their divergence from the Sorghum lineage. Subsequent Saccharum-specific autopolyploidy may have created pseudo-paralogous chromosome groups with random pairing within a group but infrequent pairing between groups. High chromosome number may reduce differentiation among Saccharum pseudo-paralogs by increasing opportunities for recombinations, with the lower chromosome numbers of Miscanthus favoring the return to disomic inheritance. The widespread tendency of plant chromosome numbers to recursively return to a narrow range following genome duplication appears to be occurring now in Saccharum spontaneum based on rich polymorphism for chromosome number among genotypes, with past reductions indicated by condensations of two ancestral chromosomes in Miscanthus (now n = 19) and perhaps as many as 10 in the Narenga-Sclerostachya clade (n = 15).
© 2014 American Society of Plant Biologists. All rights reserved.
Figures

References
-
- Al-Janabi S.M., Honeycutt R.J., Sobral B.W.S. (1994). Chromosome assortment in Saccharum. Theor. Appl. Genet. 89: 959–963. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

