Characterization of THB1, a Chlamydomonas reinhardtii truncated hemoglobin: linkage to nitrogen metabolism and identification of lysine as the distal heme ligand
- PMID: 24964018
- PMCID: PMC4108185
- DOI: 10.1021/bi5005206
Characterization of THB1, a Chlamydomonas reinhardtii truncated hemoglobin: linkage to nitrogen metabolism and identification of lysine as the distal heme ligand
Abstract
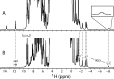
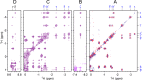
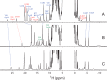
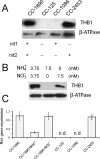
The nuclear genome of the model organism Chlamydomonas reinhardtii contains genes for a dozen hemoglobins of the truncated lineage. Of those, THB1 is known to be expressed, but the product and its function have not yet been characterized. We present mutagenesis, optical, and nuclear magnetic resonance data for the recombinant protein and show that at pH near neutral in the absence of added ligand, THB1 coordinates the heme iron with the canonical proximal histidine and a distal lysine. In the cyanomet state, THB1 is structurally similar to other known truncated hemoglobins, particularly the heme domain of Chlamydomonas eugametos LI637, a light-induced chloroplastic hemoglobin. Recombinant THB1 is capable of binding nitric oxide (NO(•)) in either the ferric or ferrous state and has efficient NO(•) dioxygenase activity. By using different C. reinhardtii strains and growth conditions, we demonstrate that the expression of THB1 is under the control of the NIT2 regulatory gene and that the hemoglobin is linked to the nitrogen assimilation pathway.
Figures





References
-
- Potts M.; Angeloni S. V.; Ebel R. E.; Bassam D. (1992) Myoglobin in a cyanobacterium. Science 256, 1690–1691. - PubMed
-
- Wittenberg J. B.; Bolognesi M.; Wittenberg B. A.; Guertin M. (2002) Truncated hemoglobins: A new family of hemoglobins widely distributed in bacteria, unicellular eukaryotes and plants. J. Biol. Chem. 277, 871–874. - PubMed
-
- Couture M.; Chamberland H.; St-Pierre B.; Lafontaine J.; Guertin M. (1994) Nuclear genes encoding chloroplast hemoglobins in the unicellular green alga Chlamydomonas eugametos. Mol. Gen. Genet. 243, 185–197. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

