Identification of endogenous reference genes for the analysis of microRNA expression in the hippocampus of the pilocarpine-induced model of mesial temporal lobe epilepsy
- PMID: 24964029
- PMCID: PMC4070922
- DOI: 10.1371/journal.pone.0100529
Identification of endogenous reference genes for the analysis of microRNA expression in the hippocampus of the pilocarpine-induced model of mesial temporal lobe epilepsy
Abstract
Real-time quantitative RT-PCR (qPCR) is one of the most powerful techniques for analyzing miRNA expression because of its sensitivity and specificity. However, in this type of analysis, a suitable normalizer is required to ensure that gene expression is unaffected by the experimental condition. To the best of our knowledge, there are no reported studies that performed a detailed identification and validation of suitable reference genes for miRNA qPCR during the epileptogenic process. Here, using a pilocarpine (PILO) model of mesial temporal lobe epilepsy (MTLE), we investigated five potential reference genes, performing a stability expression analysis using geNorm and NormFinder softwares. As a validation strategy, we used each one of the candidate reference genes to measure PILO-induced changes in microRNA-146a levels, a gene whose expression pattern variation in the PILO injected model is known. Our results indicated U6SnRNA and SnoRNA as the most stable candidate reference genes. By geNorm analysis, the normalization factor should preferably contain at least two of the best candidate reference genes (snoRNA and U6SnRNA). In fact, when normalized using the best combination of reference genes, microRNA-146a transcripts were found to be significantly increased in chronic stage, which is consistent with the pattern reported in different models. Conversely, when reference genes were individually employed for normalization, we failed to detect up-regulation of the microRNA-146a gene in the hippocampus of epileptic rats. The data presented here support that the combination of snoRNA and U6SnRNA was the minimum necessary for an accurate normalization of gene expression at the different stages of epileptogenesis that we tested.
Conflict of interest statement
Figures
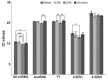
 SD (n = 6). ANOVA, *
SD (n = 6). ANOVA, *  , **
, **  , ***
, ***  .
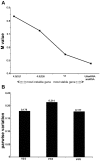
.
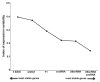
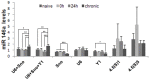
 SD, n = 6). The diagram shows mean levels of miR-146a transcripts in naive animals, epileptogenesis (0 h and 24 h) and chronic period. ANOVA, *
SD, n = 6). The diagram shows mean levels of miR-146a transcripts in naive animals, epileptogenesis (0 h and 24 h) and chronic period. ANOVA, *  , **
, **  , ***
, ***  .
.References
-
- Mathern GW, Adelson PD, Cahan LD, Leite JP (2002) Hippocampal neuron damage in human epilepsy: Meyer's hypothesis revisited. Prog Brain Res 135: 237–251. - PubMed
-
- Wiebe S, Blume WT, Girvin JP, Eliasziw M (2001) A randomized, controlled trial of surgery for temporal-lobe epilepsy. N Engl J Med 345: 311–318. - PubMed
-
- Pitkanen A, Lukasiuk K (2009) Molecular and cellular basis of epileptogenesis in symptomatic epilepsy. Epilepsy Behav 14 Suppl 116–25. - PubMed
-
- Becker AJ, Chen J, Zien A, Sochivko D, Normann S, et al. (2003) Correlated stage- and subfield-associated hippocampal gene expression patterns in experimental and human temporal lobe epilepsy. Eur J Neurosci 18: 2792–2802. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

