Cathepsin S signals via PAR2 and generates a novel tethered ligand receptor agonist
- PMID: 24964046
- PMCID: PMC4070910
- DOI: 10.1371/journal.pone.0099702
Cathepsin S signals via PAR2 and generates a novel tethered ligand receptor agonist
Abstract
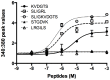
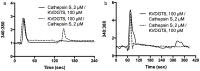
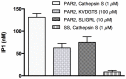
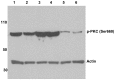
Protease-activated receptor-2 is widely expressed in mammalian epithelial, immune and neural tissues. Cleavage of PAR2 by serine proteases leads to self-activation of the receptor by the tethered ligand SLIGRL. The contribution of other classes of proteases to PAR activation has not been studied in detail. Cathepsin S is a widely expressed cysteine protease that is upregulated in inflammatory conditions. It has been suggested that cathepsin S activates PAR2. However, cathepsin S activation of PAR2 has not been demonstrated directly nor has the potential mechanism of activation been identified. We show that cathepsin S cleaves near the N-terminus of PAR2 to expose a novel tethered ligand, KVDGTS. The hexapeptide KVDGTS generates downstream signaling events specific to PAR2 but is weaker than SLIGRL. Mutation of the cathepsin S cleavage site prevents receptor activation by the protease while KVDGTS retains activity. In conclusion, the range of actions previously ascribed to cysteine cathepsins in general, and cathepsin S in particular, should be expanded to include molecular signaling. Such signaling may link together observations that had been attributed previously to PAR2 or cathepsin S individually. These interactions may contribute to inflammation.
Conflict of interest statement
Figures



References
-
- Coughlin SR (2005) Protease-activated receptors in hemostasis, thrombosis and vascular biology. J Thromb Haemost 3: 1800–1814. - PubMed
-
- Moormann C, Artuc M, Pohl E, Varga G, Buddenkotte J, et al. (2006) Functional characterization and expression analysis of the proteinase-activated receptor-2 in human cutaneous mast cells. J Invest Dermatol 126: 746–755. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

