The Trichoderma reesei Cry1 protein is a member of the cryptochrome/photolyase family with 6-4 photoproduct repair activity
- PMID: 24964051
- PMCID: PMC4070973
- DOI: 10.1371/journal.pone.0100625
The Trichoderma reesei Cry1 protein is a member of the cryptochrome/photolyase family with 6-4 photoproduct repair activity
Abstract
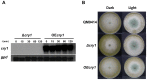
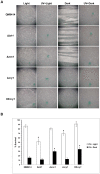
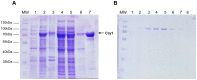
DNA-photolyases use UV-visible light to repair DNA damage caused by UV radiation. The two major types of DNA damage are cyclobutane pyrimidine dimers (CPD) and 6-4 photoproducts (6-4PP), which are repaired under illumination by CPD and 6-4 photolyases, respectively. Cryptochromes are proteins related to DNA photolyases with strongly reduced or lost DNA repair activity, and have been shown to function as blue-light photoreceptors and to play important roles in circadian rhythms in plants and animals. Both photolyases and cryptochromes belong to the cryptochrome/photolyase family, and are widely distributed in all organisms. Here we describe the characterization of cry1, a member of the cryptochrome/photolyase protein family of the filamentous fungus Trichoderma reesei. We determined that cry1 transcript accumulates when the fungus is exposed to light, and that such accumulation depends on the photoreceptor Blr1 and is modulated by Envoy. Conidia of cry1 mutants show decreased photorepair capacity of DNA damage caused by UV light. In contrast, strains over-expressing Cry1 show increased repair, as compared to the parental strain even in the dark. These observations suggest that Cry1 may be stimulating other systems involved in DNA repair, such as the nucleotide excision repair system. We show that Cry1, heterologously expressed and purified from E. coli, is capable of binding to undamaged and 6-4PP damaged DNA. Photorepair assays in vitro clearly show that Cry1 repairs 6-4PP, but not CPD and Dewar DNA lesions.
Conflict of interest statement
Figures




Similar articles
-
Residues at a Single Site Differentiate Animal Cryptochromes from Cyclobutane Pyrimidine Dimer Photolyases by Affecting the Proteins' Preferences for Reduced FAD.Chembiochem. 2017 Jun 19;18(12):1129-1137. doi: 10.1002/cbic.201700145. Epub 2017 May 15. Chembiochem. 2017. PMID: 28393477
-
The DASH-type Cryptochrome from the Fungus Mucor circinelloides Is a Canonical CPD-Photolyase.Curr Biol. 2020 Nov 16;30(22):4483-4490.e4. doi: 10.1016/j.cub.2020.08.051. Epub 2020 Sep 17. Curr Biol. 2020. PMID: 32946746
-
Trichoderma atroviride PHR1, a fungal photolyase responsible for DNA repair, autoregulates its own photoinduction.Eukaryot Cell. 2007 Sep;6(9):1682-92. doi: 10.1128/EC.00208-06. Epub 2007 Jun 1. Eukaryot Cell. 2007. PMID: 17545314 Free PMC article.
-
DNA repair by photolyases.Adv Protein Chem Struct Biol. 2019;115:1-19. doi: 10.1016/bs.apcsb.2018.10.003. Epub 2018 Dec 20. Adv Protein Chem Struct Biol. 2019. PMID: 30798929 Review.
-
Photolyase: Dynamics and electron-transfer mechanisms of DNA repair.Arch Biochem Biophys. 2017 Oct 15;632:158-174. doi: 10.1016/j.abb.2017.08.007. Epub 2017 Aug 9. Arch Biochem Biophys. 2017. PMID: 28802828 Free PMC article. Review.
Cited by
-
The Two Cryptochrome/Photolyase Family Proteins Fulfill Distinct Roles in DNA Photorepair and Regulation of Conidiation in the Gray Mold Fungus Botrytis cinerea.Appl Environ Microbiol. 2017 Aug 17;83(17):e00812-17. doi: 10.1128/AEM.00812-17. Print 2017 Sep 1. Appl Environ Microbiol. 2017. PMID: 28667107 Free PMC article.
-
The Genomes of Three Uneven Siblings: Footprints of the Lifestyles of Three Trichoderma Species.Microbiol Mol Biol Rev. 2016 Feb 10;80(1):205-327. doi: 10.1128/MMBR.00040-15. Print 2016 Mar. Microbiol Mol Biol Rev. 2016. PMID: 26864432 Free PMC article. Review.
-
A Global Analysis of Photoreceptor-Mediated Transcriptional Changes Reveals the Intricate Relationship Between Central Metabolism and DNA Repair in the Filamentous Fungus Trichoderma atroviride.Front Microbiol. 2021 Sep 8;12:724676. doi: 10.3389/fmicb.2021.724676. eCollection 2021. Front Microbiol. 2021. PMID: 34566928 Free PMC article.
-
Fungal photobiology: visible light as a signal for stress, space and time.Curr Genet. 2015 Aug;61(3):275-88. doi: 10.1007/s00294-014-0451-0. Epub 2014 Oct 17. Curr Genet. 2015. PMID: 25323429 Free PMC article. Review.
-
Two Photolyases Repair Distinct DNA Lesions and Reactivate UVB-Inactivated Conidia of an Insect Mycopathogen under Visible Light.Appl Environ Microbiol. 2019 Feb 6;85(4):e02459-18. doi: 10.1128/AEM.02459-18. Print 2019 Feb 15. Appl Environ Microbiol. 2019. PMID: 30552186 Free PMC article.
References
-
- Casas-Flores S, Herrera-Estrella A, Mukherjee P, Horwitz B, Singh U, et al. (2013) The influence of light on the biology of Trichoderma Trichoderma: biology and applications. pp. 43–66. DOI: 10.1079/9781780642475.0043
-
- Heintzen C, Loros JJ, Dunlap JC (2001) The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell 104: 453–464. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

