Exploring community structure in biological networks with random graphs
- PMID: 24965130
- PMCID: PMC4094994
- DOI: 10.1186/1471-2105-15-220
Exploring community structure in biological networks with random graphs
Abstract
Background: Community structure is ubiquitous in biological networks. There has been an increased interest in unraveling the community structure of biological systems as it may provide important insights into a system's functional components and the impact of local structures on dynamics at a global scale. Choosing an appropriate community detection algorithm to identify the community structure in an empirical network can be difficult, however, as the many algorithms available are based on a variety of cost functions and are difficult to validate. Even when community structure is identified in an empirical system, disentangling the effect of community structure from other network properties such as clustering coefficient and assortativity can be a challenge.
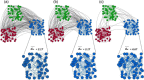
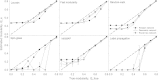
Results: Here, we develop a generative model to produce undirected, simple, connected graphs with a specified degrees and pattern of communities, while maintaining a graph structure that is as random as possible. Additionally, we demonstrate two important applications of our model: (a) to generate networks that can be used to benchmark existing and new algorithms for detecting communities in biological networks; and (b) to generate null models to serve as random controls when investigating the impact of complex network features beyond the byproduct of degree and modularity in empirical biological networks.
Conclusion: Our model allows for the systematic study of the presence of community structure and its impact on network function and dynamics. This process is a crucial step in unraveling the functional consequences of the structural properties of biological systems and uncovering the mechanisms that drive these systems.
Figures




References
-
- Newman M. Mixing patterns in networks. Phys Rev E. 2003;67(2):026126. - PubMed
-
- Ravasz E, Somera AL, Oltvai ZN, Barabási AL. Mongru Da. Hierarchical organization of modularity in metabolic networks. Science (New York, NY) 2002;297(5586):1551–1555. doi: 10.1126/science.1073374. [ http://www.ncbi.nlm.nih.gov/pubmed/12202830] - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

