Oligotyping analysis of the human oral microbiome
- PMID: 24965363
- PMCID: PMC4104879
- DOI: 10.1073/pnas.1409644111
Oligotyping analysis of the human oral microbiome
Abstract
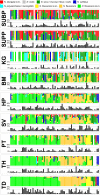
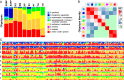
The Human Microbiome Project provided a census of bacterial populations in healthy individuals, but an understanding of the biomedical significance of this census has been hindered by limited taxonomic resolution. A high-resolution method termed oligotyping overcomes this limitation by evaluating individual nucleotide positions using Shannon entropy to identify the most information-rich nucleotide positions, which then define oligotypes. We have applied this method to comprehensively analyze the oral microbiome. Using Human Microbiome Project 16S rRNA gene sequence data for the nine sites in the oral cavity, we identified 493 oligotypes from the V1-V3 data and 360 oligotypes from the V3-V5 data. We associated these oligotypes with species-level taxon names by comparison with the Human Oral Microbiome Database. We discovered closely related oligotypes, differing sometimes by as little as a single nucleotide, that showed dramatically different distributions among oral sites and among individuals. We also detected potentially pathogenic taxa in high abundance in individual samples. Numerous oligotypes were preferentially located in plaque, others in keratinized gingiva or buccal mucosa, and some oligotypes were characteristic of habitat groupings such as throat, tonsils, tongue dorsum, hard palate, and saliva. The differing habitat distributions of closely related oligotypes suggest a level of ecological and functional biodiversity not previously recognized. We conclude that the Shannon entropy approach of oligotyping has the capacity to analyze entire microbiomes, discriminate between closely related but distinct taxa and, in combination with habitat analysis, provide deep insight into the microbial communities in health and disease.
Keywords: biogeography; microbiota; mouth.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Keijser BJF, et al. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res. 2008;87(11):1016–1020. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

