Gene-targeted microfluidic cultivation validated by isolation of a gut bacterium listed in Human Microbiome Project's Most Wanted taxa
- PMID: 24965364
- PMCID: PMC4103313
- DOI: 10.1073/pnas.1404753111
Gene-targeted microfluidic cultivation validated by isolation of a gut bacterium listed in Human Microbiome Project's Most Wanted taxa
Abstract
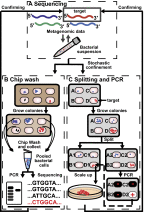
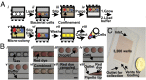
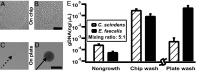
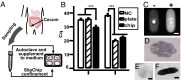
This paper describes a microfluidics-based workflow for genetically targeted isolation and cultivation of microorganisms from complex clinical samples. Data sets from high-throughput sequencing suggest the existence of previously unidentified bacterial taxa and functional genes with high biomedical importance. Obtaining isolates of these targets, preferably in pure cultures, is crucial for advancing understanding of microbial genetics and physiology and enabling physical access to microbes for further applications. However, the majority of microbes have not been cultured, due in part to the difficulties of both identifying proper growth conditions and characterizing and isolating each species. We describe a method that enables genetically targeted cultivation of microorganisms through a combination of microfluidics and on- and off-chip assays. This method involves (i) identification of cultivation conditions for microbes using growth substrates available only in small quantities as well as the correction of sampling bias using a "chip wash" technique; and (ii) performing on-chip genetic assays while also preserving live bacterial cells for subsequent scale-up cultivation of desired microbes, by applying recently developed technology to create arrays of individually addressable replica microbial cultures. We validated this targeted approach by cultivating a bacterium, here referred to as isolate microfluidicus 1, from a human cecal biopsy. Isolate microfluidicus 1 is, to our knowledge, the first successful example of targeted cultivation of a microorganism from the high-priority group of the Human Microbiome Project's "Most Wanted" list, and, to our knowledge, the first cultured representative of a previously unidentified genus of the Ruminococcaceae family.
Keywords: aerobe; anaerobe; cultivate; metagenome; microscale.
Conflict of interest statement
Conflict of interest statement: R.F.I. has a financial interest in SlipChip Corporation.
Figures


References
-
- Kennedy J, et al. Functional metagenomic strategies for the discovery of novel enzymes and biosurfactants with biotechnological applications from marine ecosystems. J Appl Microbiol. 2011;111(4):787–799. - PubMed
-
- Rooks DJ, McDonald JE, McCarthy AJ. Metagenomic approaches to the discovery of cellulases. Methods Enzymol. 2012;510:375–394. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

