The B2 flowering time locus of beet encodes a zinc finger transcription factor
- PMID: 24965366
- PMCID: PMC4104907
- DOI: 10.1073/pnas.1404829111
The B2 flowering time locus of beet encodes a zinc finger transcription factor
Abstract
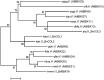
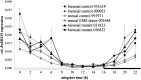
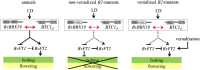
Sugar beet (Beta vulgaris) is a biennial root crop that grows vegetatively in the first year and starts shoot elongation (bolting) and flowering after exposure to cold temperatures over winter. Early bolting before winter is controlled by the dominant allele of the B locus. Recently, the BOLTING time control 1 (BTC1) gene has been cloned from this locus. BTC1 promotes early bolting through repression of the downstream bolting repressor B. vulgaris flowering locus T1 (BvFT1) and activation of the downstream floral activator BvFT2. We have identified a new bolting locus B2 acting epistatically to B. B2 houses a transcription factor which is diurnally regulated and acts like BTC1 upstream of BvFT1 and BvFT2. It was termed BvBBX19 according to its closest homolog from Arabidopsis thaliana. The encoded protein has two conserved domains with homology to zinc finger B-boxes. Ethyl methanesulfonate-induced mutations within the second B-box caused up-regulation of BvFT1 and complete down-regulation of BvFT2. In Arabidopsis, the expression of FT is promoted by the B-box containing protein CONSTANS (CO). We performed a phylogenetic analysis with B-box genes from beet and A. thaliana but only BvCOL1 clustered with CO. However, BvCOL1 had been excluded as a CO ortholog by previous studies. Therefore, a new model for flowering induction in beet is proposed in which BTC1 and BvBBX19 complement each other and thus acquire a CO function to regulate their downstream targets BvFT1 and BvFT2.
Keywords: map-based cloning; sucrose; winter beet.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13(9):627–639. - PubMed
-
- Pin PA, et al. The role of a pseudo-response regulator gene in life cycle adaptation and domestication of beet. Curr Biol. 2012;22(12):1095–1101. - PubMed
-
- Hohmann U, Jacobs G, Jung C. An EMS mutagenesis protocol for sugar beet and isolation of non-bolting mutants. Plant Breed. 2005;124(4):317–321.
-
- Abou-Elwafa S, Büttner B, Kopisch-Obuch F, Jung C, Müller AE. Genetic identification of a novel bolting locus in Beta vulgaris which promotes annuality independently of the bolting gene B. Mol Breed. 2012;29(4):989–998.
-
- Büttner B, Abou-Elwafa SF, Zhang W, Jung C, Müller AE. A survey of EMS-induced biennial Beta vulgaris mutants reveals a novel bolting locus which is unlinked to the bolting gene B. Theor Appl Genet. 2010;121(6):1117–1131. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

