Structure of 2G12 Fab2 in complex with soluble and fully glycosylated HIV-1 Env by negative-stain single-particle electron microscopy
- PMID: 24965454
- PMCID: PMC4136306
- DOI: 10.1128/JVI.01229-14
Structure of 2G12 Fab2 in complex with soluble and fully glycosylated HIV-1 Env by negative-stain single-particle electron microscopy
Abstract
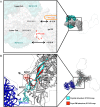
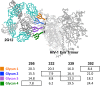
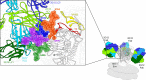
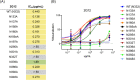
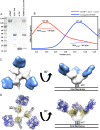
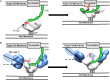
The neutralizing anti-HIV-1 antibody 2G12 is of particular interest due to the sterilizing protection it provides from viral challenge in animal models. 2G12 is a unique, domain-exchanged antibody that binds exclusively to conserved N-linked glycans that form the high-mannose patch on the gp120 outer domain centered on a glycan at position N332. Several glycans in and around the 2G12 epitope have been shown to interact with other potent, broadly neutralizing antibodies; therefore, this region constitutes a supersite of vulnerability on gp120. While crystal structures of 2G12 and 2G12 bound to high-mannose glycans have been solved, no structural information that describes the interaction of 2G12 with gp120 or the Env trimer is available. Here, we present a negative-stain single-particle electron microscopy reconstruction of 2G12 Fab2 in complex with a soluble, trimeric Env at ∼17-Å resolution that reveals the antibody's interaction with its native and fully glycosylated epitope. We also mapped relevant glycans in this epitope by fitting high-resolution crystal structures and by performing neutralization assays of glycan knockouts. In addition, a reconstruction at ∼26 Å of the ternary complex formed by 2G12 Fab2, soluble CD4, and Env indicates that 2G12 may block membrane fusion by induced steric hindrance upon primary receptor binding, thereby abrogating Env's interaction with coreceptor(s). These structures provide a basis for understanding 2G12 binding and neutralization, and our low-resolution model and glycan assignments provide a basis for higher-resolution studies to determine the molecular nature of the 2G12 epitope.
Importance: HIV-1 is a human virus that results in the deaths of millions of people around the world each year. While there are several effective therapeutics available to prolong life, a vaccine is the best long-term solution for curbing this global epidemic. Here, we present structural data that reveal the viral binding site of one of the first HIV-1-neutralizing antibodies isolated, 2G12, and provide a rationale for its effectiveness. These structures provide a basis for higher-resolution studies to determine the molecular nature of the 2G12 epitope, which will aid in vaccine design and antibody-based therapies.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- McCaffrey RA, Saunders C, Hensel M, Stamatatos L. 2004. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J. Virol. 78:3279–3295. 10.1128/JVI.78.7.3279-3295.2004 - DOI - PMC - PubMed
-
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, Phogat A, Chakrabarti B, Li Y, Connors M, Pereyra F, Walker BD, Wardemann H, Ho D, Wyatt RT, Mascola JR, Ravetch JV, Nussenzweig MC. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636–640. 10.1038/nature07930 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

