ROCC, a conserved region in cohesin's Mcd1 subunit, is essential for the proper regulation of the maintenance of cohesion and establishment of condensation
- PMID: 24966169
- PMCID: PMC4142609
- DOI: 10.1091/mbc.E14-04-0929
ROCC, a conserved region in cohesin's Mcd1 subunit, is essential for the proper regulation of the maintenance of cohesion and establishment of condensation
Abstract
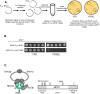
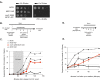
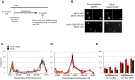
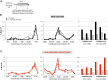
Cohesin helps orchestrate higher-order chromosome structure, thereby promoting sister chromatid cohesion, chromosome condensation, DNA repair, and transcriptional regulation. To elucidate how cohesin facilitates these diverse processes, we mutagenized Mcd1p, the kleisin regulatory subunit of budding yeast cohesin. In the linker region of Mcd1p, we identified a novel evolutionarily conserved 10-amino acid cluster, termed the regulation of cohesion and condensation (ROCC) box. We show that ROCC promotes cohesion maintenance by protecting a second activity of cohesin that is distinct from its stable binding to chromosomes. The existence of this second activity is incompatible with the simple embrace mechanism of cohesion. In addition, we show that the ROCC box is required for the establishment of condensation. We provide evidence that ROCC controls cohesion maintenance and condensation establishment through differential functional interactions with Pds5p and Wpl1p.
© 2014 Eng et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures

References
-
- Baldwin ML, Julius JA, Tang X, Wang Y, Bachant J. The yeast SUMO isopeptidase Smt4/Ulp2 and the polo kinase Cdc5 act in an opposing fashion to regulate sumoylation in mitosis and cohesion at centromeres. Cell Cycle. 2009;8:3406–3419. - PubMed
-
- D'Ambrosio LM, Lavoie BD. Pds5 Prevents the polySUMO-dependent separation of sister chromatids. Curr Biol. 2014;24:361–371. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

