Spy: a new group of eukaryotic DNA transposons without target site duplications
- PMID: 24966181
- PMCID: PMC4122938
- DOI: 10.1093/gbe/evu140
Spy: a new group of eukaryotic DNA transposons without target site duplications
Abstract
Class 2 or DNA transposons populate the genomes of most eukaryotes and like other mobile genetic elements have a profound impact on genome evolution. Most DNA transposons belong to the cut-and-paste types, which are relatively simple elements characterized by terminal-inverted repeats (TIRs) flanking a single gene encoding a transposase. All eukaryotic cut-and-paste transposons so far described are also characterized by target site duplications (TSDs) of host DNA generated upon chromosomal insertion. Here, we report a new group of evolutionarily related DNA transposons called Spy, which also include TIRs and DDE motif-containing transposase but surprisingly do not create TSDs upon insertion. Instead, Spy transposons appear to transpose precisely between 5'-AAA and TTT-3' host nucleotides, without duplication or modification of the AAATTT target sites. Spy transposons were identified in the genomes of diverse invertebrate species based on transposase homology searches and structure-based approaches. Phylogenetic analyses indicate that Spy transposases are distantly related to IS5, ISL2EU, and PIF/Harbinger transposases. However, Spy transposons are distinct from these and other DNA transposon superfamilies by their lack of TSD and their target site preference. Our findings expand the known diversity of DNA transposons and reveal a new group of eukaryotic DDE transposases with unusual catalytic properties.
Keywords: DNA transposon; Spy; target site duplication; transposition.
© The Author(s) 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

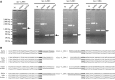
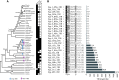


References
-
- Bao WD, Jurka J. DNA transposons from the Pacific oyster genome. Repbase Rep. 2013;13:578–580.
-
- Chandler M, Mahillon J. Washington (DC): American Society for Microbiology; 2002. Insertion sequences revisited, in mobile DNA II.
-
- Craig NL, Craigie R, Gellert M, Lambowitz AM. Washington (DC): American Society for Microbiology Press; 2002. Mobile DNA II.
-
- Craigie R, Mizuuchi K. Mechanism of transposition of bacteriophage Mu: structure of a transposition intermediate. Cell. 1985;41:867–876. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

