An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases
- PMID: 24966208
- PMCID: PMC4109310
- DOI: 10.7554/eLife.02304
An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases
Abstract
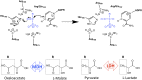
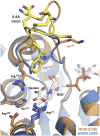
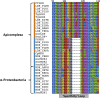
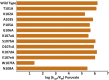
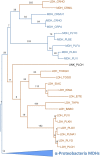
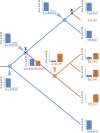
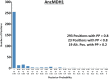
Malate and lactate dehydrogenases (MDH and LDH) are homologous, core metabolic enzymes that share a fold and catalytic mechanism yet possess strict specificity for their substrates. In the Apicomplexa, convergent evolution of an unusual LDH from MDH produced a difference in specificity exceeding 12 orders of magnitude. The mechanisms responsible for this extraordinary functional shift are currently unknown. Using ancestral protein resurrection, we find that specificity evolved in apicomplexan LDHs by classic neofunctionalization characterized by long-range epistasis, a promiscuous intermediate, and few gain-of-function mutations of large effect. In canonical MDHs and LDHs, a single residue in the active-site loop governs substrate specificity: Arg102 in MDHs and Gln102 in LDHs. During the evolution of the apicomplexan LDH, however, specificity switched via an insertion that shifted the position and identity of this 'specificity residue' to Trp107f. Residues far from the active site also determine specificity, as shown by the crystal structures of three ancestral proteins bracketing the key duplication event. This work provides an unprecedented atomic-resolution view of evolutionary trajectories creating a nascent enzymatic function.
Keywords: ancestral sequence reconstruction; biophysics; dehydrogenase; evolutionary biology; gene duplication; genomics; molecular evolution; promiscuity; specificity; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
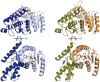










References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography 66:213–221. doi: 10.1107/S0907444909052925 - DOI - PMC - PubMed
-
- Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, Heiges M, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Miller JA, Nayak V, Pennington C, Pinney DF, Roos DS, Ross C, Stoeckert CJ, Jnr, Treatman C, Wang H. 2009. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Research 37:D539–D543. doi: 10.1093/nar/gkn814 - DOI - PMC - PubMed
-
- Birktoft JJ, Banaszak LJ. 1983. The presence of a histidine-aspartic acid pair in the active site of 2-hydroxyacid dehydrogenases. X-ray refinement of cytoplasmic malate dehydrogenase. The Journal of Biological Chemistry 258:472–482 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

