Transition-state destabilization reveals how human DNA polymerase β proceeds across the chemically unstable lesion N7-methylguanine
- PMID: 24966350
- PMCID: PMC4117778
- DOI: 10.1093/nar/gku554
Transition-state destabilization reveals how human DNA polymerase β proceeds across the chemically unstable lesion N7-methylguanine
Abstract
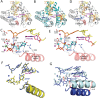
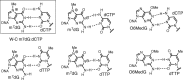
N7-Methyl-2'-deoxyguanosine (m7dG) is the predominant lesion formed by methylating agents. A systematic investigation on the effect of m7dG on DNA replication has been difficult due to the chemical instability of m7dG. To gain insights into the m7dG effect, we employed a 2'-fluorine-mediated transition-state destabilzation strategy. Specifically, we determined kinetic parameters for dCTP insertion opposite a chemically stable m7dG analogue, 2'-fluoro-m7dG (Fm7dG), by human DNA polymerase β (polβ) and solved three X-ray structures of polβ in complex with the templating Fm7dG paired with incoming dCTP or dTTP analogues. The kinetic studies reveal that the templating Fm7dG slows polβ catalysis ∼ 300-fold, suggesting that m7dG in genomic DNA may impede replication by some DNA polymerases. The structural analysis reveals that Fm7dG forms a canonical Watson-Crick base pair with dCTP, but metal ion coordination is suboptimal for catalysis in the polβ-Fm7dG:dCTP complex, which partially explains the slow insertion of dCTP opposite Fm7dG by polβ. In addition, the polβ-Fm7dG:dTTP structure shows open protein conformations and staggered base pair conformations, indicating that N7-methylation of dG does not promote a promutagenic replication. Overall, the first systematic studies on the effect of m7dG on DNA replication reveal that polβ catalysis across m7dG is slow, yet highly accurate.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
-
- Sedgwick B. Repairing DNA-methylation damage. Nat. Rev. Mol. Cell Biol. 2004;5:148–157. - PubMed
-
- Reiner B., Zamenhof S. Studies on the chemically reactive groups of deoxynucleic acids. J. Biol. Chem. 1957;228:475–486. - PubMed
-
- Hayashibara K.C., Verdine G.L. Template-directed interference footprinting of cytosine contacts in a protein-DNA complex: potent interference by 5-aza-2’-deoxycytidine. Biochemistry. 1992;31:11265–11273. - PubMed
-
- Ezaz-Nikpay K., Verdine G.L. Aberrantly methylated DNA: site-specific introduction of N-7-methyl-2’-deoxyguanosine into the Dickerson/Drew dodecamer. J. Am. Chem. Soc. 1992;114:6562–6563.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

