Fructose-asparagine is a primary nutrient during growth of Salmonella in the inflamed intestine
- PMID: 24967579
- PMCID: PMC4072780
- DOI: 10.1371/journal.ppat.1004209
Fructose-asparagine is a primary nutrient during growth of Salmonella in the inflamed intestine
Abstract
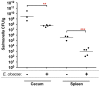
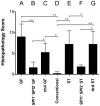
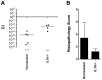
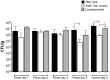
Salmonella enterica serovar Typhimurium (Salmonella) is one of the most significant food-borne pathogens affecting both humans and agriculture. We have determined that Salmonella encodes an uptake and utilization pathway specific for a novel nutrient, fructose-asparagine (F-Asn), which is essential for Salmonella fitness in the inflamed intestine (modeled using germ-free, streptomycin-treated, ex-germ-free with human microbiota, and IL10-/- mice). The locus encoding F-Asn utilization, fra, provides an advantage only if Salmonella can initiate inflammation and use tetrathionate as a terminal electron acceptor for anaerobic respiration (the fra phenotype is lost in Salmonella SPI1- SPI2- or ttrA mutants, respectively). The severe fitness defect of a Salmonella fra mutant suggests that F-Asn is the primary nutrient utilized by Salmonella in the inflamed intestine and that this system provides a valuable target for novel therapies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

