Reviving the dead: history and reactivation of an extinct l1
- PMID: 24968166
- PMCID: PMC4072516
- DOI: 10.1371/journal.pgen.1004395
Reviving the dead: history and reactivation of an extinct l1
Abstract
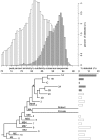
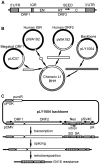
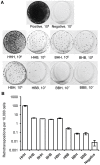
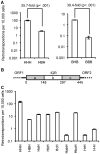
Although L1 sequences are present in the genomes of all placental mammals and marsupials examined to date, their activity was lost in the megabat family, Pteropodidae, ∼24 million years ago. To examine the characteristics of L1s prior to their extinction, we analyzed the evolutionary history of L1s in the genome of a megabat, Pteropus vampyrus, and found a pattern of periodic L1 expansion and quiescence. In contrast to the well-characterized L1s in human and mouse, megabat genomes have accommodated two or more simultaneously active L1 families throughout their evolutionary history, and major peaks of L1 deposition into the genome always involved multiple families. We compared the consensus sequences of the two major megabat L1 families at the time of their extinction to consensus L1s of a variety of mammalian species. Megabat L1s are comparable to the other mammalian L1s in terms of adenosine content and conserved amino acids in the open reading frames (ORFs). However, the intergenic region (IGR) of the reconstructed element from the more active family is dramatically longer than the IGR of well-characterized human and mouse L1s. We synthesized the reconstructed element from this L1 family and tested the ability of its components to support retrotransposition in a tissue culture assay. Both ORFs are capable of supporting retrotransposition, while the IGR is inhibitory to retrotransposition, especially when combined with either of the reconstructed ORFs. We dissected the inhibitory effect of the IGR by testing truncated and shuffled versions and found that length is a key factor, but not the only one affecting inhibition of retrotransposition. Although the IGR is inhibitory to retrotransposition, this inhibition does not account for the extinction of L1s in megabats. Overall, the evolution of the L1 sequence or the quiescence of L1 is unlikely the reason of L1 extinction.
Conflict of interest statement
The authors have no competing interests.
Figures

References
-
- Furano AV (2000) The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog Nucleic Acid Res Mol Biol 64: 255–294. - PubMed
-
- Kulpa DA, Moran JV (2006) Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol 13: 655–660. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

