Genome downsizing and karyotype constancy in diploid and polyploid congeners: a model of genome size variation
- PMID: 24969503
- PMCID: PMC4152747
- DOI: 10.1093/aobpla/plu029
Genome downsizing and karyotype constancy in diploid and polyploid congeners: a model of genome size variation
Abstract
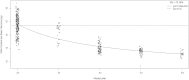
Evolutionary chromosome change involves significant variation in DNA amount in diploids and genome downsizing in polyploids. Genome size and karyotype parameters of Hippeastrum species with different ploidy level were analysed. In Hippeastrum, polyploid species show less DNA content per basic genome than diploid species. The rate of variation is lower at higher ploidy levels. All the species have a basic number x = 11 and bimodal karyotypes. The basic karyotypes consist of four short metacentric chromosomes and seven large chromosomes (submetacentric and subtelocentric). The bimodal karyotype is preserved maintaining the relative proportions of members of the haploid chromosome set, even in the presence of genome downsizing. The constancy of the karyotype is maintained because changes in DNA amount are proportional to the length of the whole-chromosome complement and vary independently in the long and short sets of chromosomes. This karyotype constancy in taxa of Hippeastrum with different genome size and ploidy level indicates that the distribution of extra DNA within the complement is not at random and suggests the presence of mechanisms selecting for constancy, or against changes, in karyotype morphology.
Keywords: Bimodal karyotype; DNA amount variation; Hippeastrum; genome size; karyotype constancy; polyploids..
Published by Oxford University Press on behalf of the Annals of Botany Company.
Figures



Similar articles
-
Karyotype and genome size variation in white-flowered Eranthis sect. Shibateranthis (Ranunculaceae).PhytoKeys. 2021 Dec 31;187:207-227. doi: 10.3897/phytokeys.187.75715. eCollection 2021. PhytoKeys. 2021. PMID: 35068976 Free PMC article.
-
New estimates and synthesis of chromosome numbers, ploidy levels and genome size variation in Allium sect. Codonoprasum: advancing our understanding of the unresolved diversification and evolution of this section.Bot Stud. 2024 Dec 24;65(1):40. doi: 10.1186/s40529-024-00446-8. Bot Stud. 2024. PMID: 39718713 Free PMC article.
-
Karyotype and genome size variation in Delphinium subg. Anthriscifolium (Ranunculaceae).PhytoKeys. 2023 Oct 18;234:145-165. doi: 10.3897/phytokeys.234.108841. eCollection 2023. PhytoKeys. 2023. PMID: 37901134 Free PMC article.
-
Genetic and genomic interactions of animals with different ploidy levels.Cytogenet Genome Res. 2013;140(2-4):117-36. doi: 10.1159/000351593. Epub 2013 Jun 8. Cytogenet Genome Res. 2013. PMID: 23751376 Review.
-
Tracing the Evolution of the Angiosperm Genome from the Cytogenetic Point of View.Plants (Basel). 2022 Mar 16;11(6):784. doi: 10.3390/plants11060784. Plants (Basel). 2022. PMID: 35336666 Free PMC article. Review.
Cited by
-
Exploring karyotype diversity of Argentinian Guaraní maize landraces: Relationship among South American maize.PLoS One. 2018 Jun 7;13(6):e0198398. doi: 10.1371/journal.pone.0198398. eCollection 2018. PLoS One. 2018. PMID: 29879173 Free PMC article.
-
Evolutionary importance of the relationship between cytogeography and climate: New insights on creosote bushes from North and South America.Plant Divers. 2021 Nov 30;44(5):492-498. doi: 10.1016/j.pld.2021.11.006. eCollection 2022 Sep. Plant Divers. 2021. PMID: 36187552 Free PMC article.
-
Molecular and Cytogenetic Study of East African Highland Banana.Front Plant Sci. 2018 Oct 4;9:1371. doi: 10.3389/fpls.2018.01371. eCollection 2018. Front Plant Sci. 2018. PMID: 30337933 Free PMC article.
-
Intra-specific variation in genome size in maize: cytological and phenotypic correlates.AoB Plants. 2015 Dec 7;8:plv138. doi: 10.1093/aobpla/plv138. AoB Plants. 2015. PMID: 26644343 Free PMC article.
-
Differential Repeat Accumulation in the Bimodal Karyotype of Agave L.Genes (Basel). 2023 Feb 15;14(2):491. doi: 10.3390/genes14020491. Genes (Basel). 2023. PMID: 36833420 Free PMC article.
References
-
- Aitken AC. On least squares and linear combinations of observations. Proceedings of the Royal Society of Edinburgh. 1935;55:42–48.
-
- Arroyo SC. The chromosomes of Hippeastrum, Amaryllis and Phycella (Amaryllidaceae) Kew Bulletin. 1982;37:211–216.
-
- Beltrao GT, Guerra M. Cytogenetica de angiospermas coletadas em Pernambuco. Ciência e Cultura. 1990;42:839–845.
-
- Bennett MD, Smith JB. Nuclear DNA amounts in angiosperms. Proceedings of the Royal Society of London. 1976;274:227–274. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

