Microbial evolution. Global epistasis makes adaptation predictable despite sequence-level stochasticity
- PMID: 24970088
- PMCID: PMC4314286
- DOI: 10.1126/science.1250939
Microbial evolution. Global epistasis makes adaptation predictable despite sequence-level stochasticity
Abstract
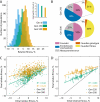
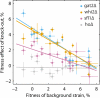
Epistatic interactions between mutations can make evolutionary trajectories contingent on the chance occurrence of initial mutations. We used experimental evolution in Saccharomyces cerevisiae to quantify this contingency, finding differences in adaptability among 64 closely related genotypes. Despite these differences, sequencing of 104 evolved clones showed that initial genotype did not constrain future mutational trajectories. Instead, reconstructed combinations of mutations revealed a pattern of diminishing-returns epistasis: Beneficial mutations have consistently smaller effects in fitter backgrounds. Taken together, these results show that beneficial mutations affecting a variety of biological processes are globally coupled; they interact strongly, but only through their combined effect on fitness. As a consequence, fitness evolution follows a predictable trajectory even though sequence-level adaptation is stochastic.
Copyright © 2014, American Association for the Advancement of Science.
Figures
Similar articles
-
Changes in the distribution of fitness effects and adaptive mutational spectra following a single first step towards adaptation.Nat Commun. 2021 Aug 31;12(1):5193. doi: 10.1038/s41467-021-25440-7. Nat Commun. 2021. PMID: 34465770 Free PMC article.
-
Modular epistasis and the compensatory evolution of gene deletion mutants.PLoS Genet. 2019 Feb 15;15(2):e1007958. doi: 10.1371/journal.pgen.1007958. eCollection 2019 Feb. PLoS Genet. 2019. PMID: 30768593 Free PMC article.
-
Negative epistasis between beneficial mutations in an evolving bacterial population.Science. 2011 Jun 3;332(6034):1193-6. doi: 10.1126/science.1203801. Science. 2011. PMID: 21636772
-
Genomic investigations of evolutionary dynamics and epistasis in microbial evolution experiments.Curr Opin Genet Dev. 2015 Dec;35:33-9. doi: 10.1016/j.gde.2015.08.008. Epub 2015 Sep 14. Curr Opin Genet Dev. 2015. PMID: 26370471 Free PMC article. Review.
-
Negative Epistasis in Experimental RNA Fitness Landscapes.J Mol Evol. 2017 Dec;85(5-6):159-168. doi: 10.1007/s00239-017-9817-5. Epub 2017 Nov 10. J Mol Evol. 2017. PMID: 29127445 Review.
Cited by
-
How microscopic epistasis and clonal interference shape the fitness trajectory in a spin glass model of microbial long-term evolution.Elife. 2024 Feb 20;12:RP87895. doi: 10.7554/eLife.87895. Elife. 2024. PMID: 38376390 Free PMC article.
-
Sulfur isotope fractionation during the evolutionary adaptation of a sulfate-reducing bacterium.Appl Environ Microbiol. 2015 Apr;81(8):2676-89. doi: 10.1128/AEM.03476-14. Epub 2015 Feb 6. Appl Environ Microbiol. 2015. PMID: 25662968 Free PMC article.
-
The evolutionarily stable distribution of fitness effects.Genetics. 2015 May;200(1):321-9. doi: 10.1534/genetics.114.173815. Epub 2015 Mar 10. Genetics. 2015. PMID: 25762525 Free PMC article.
-
Exploring a Local Genetic Interaction Network Using Evolutionary Replay Experiments.Mol Biol Evol. 2021 Jul 29;38(8):3144-3152. doi: 10.1093/molbev/msab087. Mol Biol Evol. 2021. PMID: 33749796 Free PMC article.
-
The Adaptive Potential of the Middle Domain of Yeast Hsp90.Mol Biol Evol. 2021 Jan 23;38(2):368-379. doi: 10.1093/molbev/msaa211. Mol Biol Evol. 2021. PMID: 32871012 Free PMC article.
References
-
- Burch CL, Chao L. Evolvability of an RNA virus is determined by its mutational neighbourhood. Nature. 2000;406:625. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

