Molecular Insights into Poly(ADP-ribose) Recognition and Processing
- PMID: 24970154
- PMCID: PMC4030884
- DOI: 10.3390/biom3010001
Molecular Insights into Poly(ADP-ribose) Recognition and Processing
Abstract
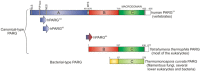
Poly(ADP-ribosyl)ation is a post-translational protein modification involved in the regulation of important cellular functions including DNA repair, transcription, mitosis and apoptosis. The amount of poly(ADP-ribosyl)ation (PAR) in cells reflects the balance of synthesis, mediated by the PARP protein family, and degradation, which is catalyzed by a glycohydrolase, PARG. Many of the proteins mediating PAR metabolism possess specialised high affinity PAR-binding modules that allow the efficient sensing or processing of the PAR signal. The identification of four such PAR-binding modules and the characterization of a number of proteins utilising these elements during the last decade has provided important insights into how PAR regulates different cellular activities. The macrodomain represents a unique PAR-binding module which is, in some instances, known to possess enzymatic activity on ADP-ribose derivatives (in addition to PAR-binding). The most recently discovered example for this is the PARG protein, and several available PARG structures have provided an understanding into how the PARG macrodomain evolved into a major enzyme that maintains PAR homeostasis in living cells.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

