Quantum mechanical modeling: a tool for the understanding of enzyme reactions
- PMID: 24970187
- PMCID: PMC4030948
- DOI: 10.3390/biom3030662
Quantum mechanical modeling: a tool for the understanding of enzyme reactions
Abstract
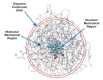
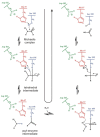
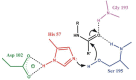
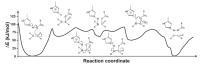
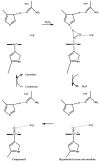
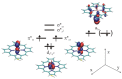
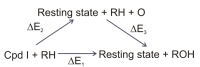
Most enzyme reactions involve formation and cleavage of covalent bonds, while electrostatic effects, as well as dynamics of the active site and surrounding protein regions, may also be crucial. Accordingly, special computational methods are needed to provide an adequate description, which combine quantum mechanics for the reactive region with molecular mechanics and molecular dynamics describing the environment and dynamic effects, respectively. In this review we intend to give an overview to non-specialists on various enzyme models as well as established computational methods and describe applications to some specific cases. For the treatment of various enzyme mechanisms, special approaches are often needed to obtain results, which adequately refer to experimental data. As a result of the spectacular progress in the last two decades, most enzyme reactions can be quite precisely treated by various computational methods.
Figures
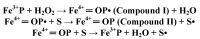

Similar articles
-
Hybrid schemes based on quantum mechanics/molecular mechanics simulations goals to success, problems, and perspectives.Adv Protein Chem Struct Biol. 2011;85:81-142. doi: 10.1016/B978-0-12-386485-7.00003-X. Adv Protein Chem Struct Biol. 2011. PMID: 21920322 Review.
-
Atomistic insight into the catalytic mechanism of glycosyltransferases by combined quantum mechanics/molecular mechanics (QM/MM) methods.Carbohydr Res. 2015 Feb 11;403:38-47. doi: 10.1016/j.carres.2014.06.017. Epub 2014 Jun 24. Carbohydr Res. 2015. PMID: 25060837 Review.
-
Combined quantum mechanics/molecular mechanics (QM/MM) methods in computational enzymology.Biochemistry. 2013 Apr 23;52(16):2708-28. doi: 10.1021/bi400215w. Epub 2013 Apr 12. Biochemistry. 2013. PMID: 23557014 Review.
-
Theoretical modeling of large molecular systems. Advances in the local self consistent field method for mixed quantum mechanics/molecular mechanics calculations.Acc Chem Res. 2013 Feb 19;46(2):596-603. doi: 10.1021/ar300278j. Epub 2012 Dec 18. Acc Chem Res. 2013. PMID: 23249409
-
Modeling biotransformation reactions by combined quantum mechanical/molecular mechanical approaches: from structure to activity.Curr Top Med Chem. 2003;3(11):1241-56. doi: 10.2174/1568026033452005. Curr Top Med Chem. 2003. PMID: 12769703 Review.
Cited by
-
Exploring Bacillus species xylanases for industrial applications: screening via thermostability and reaction modelling.J Mol Model. 2024 Jul 2;30(8):242. doi: 10.1007/s00894-024-06048-2. J Mol Model. 2024. PMID: 38955857
-
Structural Predictive Model of Presenilin-2 Protein and Analysis of Structural Effects of Familial Alzheimer's Disease Mutations.Biochem Res Int. 2021 Nov 29;2021:9542038. doi: 10.1155/2021/9542038. eCollection 2021. Biochem Res Int. 2021. PMID: 34881055 Free PMC article.
-
Complement component factor B has thrombin-like activity.Biochem Biophys Res Commun. 2021 May 7;552:17-22. doi: 10.1016/j.bbrc.2021.02.134. Epub 2021 Mar 16. Biochem Biophys Res Commun. 2021. PMID: 33740660 Free PMC article.
-
Decoding Drug Discovery: Exploring A-to-Z In Silico Methods for Beginners.Appl Biochem Biotechnol. 2025 Mar;197(3):1453-1503. doi: 10.1007/s12010-024-05110-2. Epub 2024 Dec 4. Appl Biochem Biotechnol. 2025. PMID: 39630336 Review.
-
An investigation into the applicability of the semiempirical method PM7 for modeling the catalytic mechanism in the enzyme chymotrypsin.J Mol Model. 2017 May;23(5):154. doi: 10.1007/s00894-017-3326-8. Epub 2017 Apr 4. J Mol Model. 2017. PMID: 28378242 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

