Research applications of proteolytic enzymes in molecular biology
- PMID: 24970197
- PMCID: PMC4030975
- DOI: 10.3390/biom3040923
Research applications of proteolytic enzymes in molecular biology
Abstract
Proteolytic enzymes (also termed peptidases, proteases and proteinases) are capable of hydrolyzing peptide bonds in proteins. They can be found in all living organisms, from viruses to animals and humans. Proteolytic enzymes have great medical and pharmaceutical importance due to their key role in biological processes and in the life-cycle of many pathogens. Proteases are extensively applied enzymes in several sectors of industry and biotechnology, furthermore, numerous research applications require their use, including production of Klenow fragments, peptide synthesis, digestion of unwanted proteins during nucleic acid purification, cell culturing and tissue dissociation, preparation of recombinant antibody fragments for research, diagnostics and therapy, exploration of the structure-function relationships by structural studies, removal of affinity tags from fusion proteins in recombinant protein techniques, peptide sequencing and proteolytic digestion of proteins in proteomics. The aim of this paper is to review the molecular biological aspects of proteolytic enzymes and summarize their applications in the life sciences.
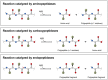
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources

