LUMPY: a probabilistic framework for structural variant discovery
- PMID: 24970577
- PMCID: PMC4197822
- DOI: 10.1186/gb-2014-15-6-r84
LUMPY: a probabilistic framework for structural variant discovery
Abstract
Comprehensive discovery of structural variation (SV) from whole genome sequencing data requires multiple detection signals including read-pair, split-read, read-depth and prior knowledge. Owing to technical challenges, extant SV discovery algorithms either use one signal in isolation, or at best use two sequentially. We present LUMPY, a novel SV discovery framework that naturally integrates multiple SV signals jointly across multiple samples. We show that LUMPY yields improved sensitivity, especially when SV signal is reduced owing to either low coverage data or low intra-sample variant allele frequency. We also report a set of 4,564 validated breakpoints from the NA12878 human genome. https://github.com/arq5x/lumpy-sv.
Figures



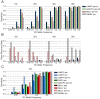


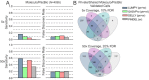

References
-
- Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat Methods. 2009;6:S13–S20. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

