Integrated RNA and DNA sequencing improves mutation detection in low purity tumors
- PMID: 24970867
- PMCID: PMC4117748
- DOI: 10.1093/nar/gku489
Integrated RNA and DNA sequencing improves mutation detection in low purity tumors
Abstract
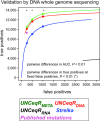
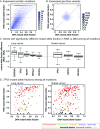
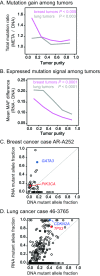
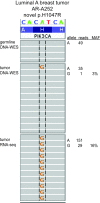
Identifying somatic mutations is critical for cancer genome characterization and for prioritizing patient treatment. DNA whole exome sequencing (DNA-WES) is currently the most popular technology; however, this yields low sensitivity in low purity tumors. RNA sequencing (RNA-seq) covers the expressed exome with depth proportional to expression. We hypothesized that integrating DNA-WES and RNA-seq would enable superior mutation detection versus DNA-WES alone. We developed a first-of-its-kind method, called UNCeqR, that detects somatic mutations by integrating patient-matched RNA-seq and DNA-WES. In simulation, the integrated DNA and RNA model outperformed the DNA-WES only model. Validation by patient-matched whole genome sequencing demonstrated superior performance of the integrated model over DNA-WES only models, including a published method and published mutation profiles. Genome-wide mutational analysis of breast and lung cancer cohorts (n = 871) revealed remarkable tumor genomics properties. Low purity tumors experienced the largest gains in mutation detection by integrating RNA-seq and DNA-WES. RNA provided greater mutation signal than DNA in expressed mutations. Compared to earlier studies on this cohort, UNCeqR increased mutation rates of driver and therapeutically targeted genes (e.g. PIK3CA, ERBB2 and FGFR2). In summary, integrating RNA-seq with DNA-WES increases mutation detection performance, especially for low purity tumors.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Korf B.R., Rehm H.L. New approaches to molecular diagnosis. JAMA. 2013;309:1511–1521. - PubMed
-
- Wilkerson M.D., Yin X., Walter V., Zhao N., Cabanski C.R., Hayward M.C., Miller C.R., Socinski M.A., Parsons A.M., Thorne L.B., et al. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PLoS One. 2012;7:e36530. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

