Sequence and expression analyses of ethylene response factors highly expressed in latex cells from Hevea brasiliensis
- PMID: 24971876
- PMCID: PMC4074046
- DOI: 10.1371/journal.pone.0099367
Sequence and expression analyses of ethylene response factors highly expressed in latex cells from Hevea brasiliensis
Abstract
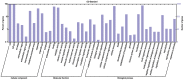
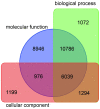
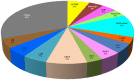
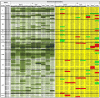
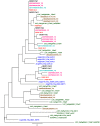
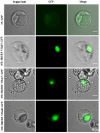
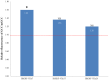
The AP2/ERF superfamily encodes transcription factors that play a key role in plant development and responses to abiotic and biotic stress. In Hevea brasiliensis, ERF genes have been identified by RNA sequencing. This study set out to validate the number of HbERF genes, and identify ERF genes involved in the regulation of latex cell metabolism. A comprehensive Hevea transcriptome was improved using additional RNA reads from reproductive tissues. Newly assembled contigs were annotated in the Gene Ontology database and were assigned to 3 main categories. The AP2/ERF superfamily is the third most represented compared with other transcription factor families. A comparison with genomic scaffolds led to an estimation of 114 AP2/ERF genes and 1 soloist in Hevea brasiliensis. Based on a phylogenetic analysis, functions were predicted for 26 HbERF genes. A relative transcript abundance analysis was performed by real-time RT-PCR in various tissues. Transcripts of ERFs from group I and VIII were very abundant in all tissues while those of group VII were highly accumulated in latex cells. Seven of the thirty-five ERF expression marker genes were highly expressed in latex. Subcellular localization and transactivation analyses suggested that HbERF-VII candidate genes encoded functional transcription factors.
Conflict of interest statement
Figures



References
-
- Latchman DS (1997) Transcription factors: An overview. The International Journal of Biochemistry & Cell Biology 29: 1305–1312. - PubMed
-
- Bleecker AB, Kende H (2000) Ethylene: a gaseous signal molecule in plants. Annu Rev Cell Dev Biol 16: 1–18. - PubMed
-
- Johnson PR, Ecker JR (1998) The ethylene gas signal transduction pathway: a molecular perspective. Annu Rev Genet 32: 227–254. - PubMed
-
- Pirrello J, Jaimes-Miranda F, Sanchez-Ballesta MT, Tournier B, Khalil-Ahmad Q, et al. (2006) Sl-ERF2, a tomato ethylene response factor involved in ethylene response and seed germination. Plant Cell Physiol 47: 1195–1205. - PubMed
-
- Dietz K-J, Vogel M, Viehhauser A (2010) AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 245: 3–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

