Phylogenomic analysis of "red" genes from two divergent species of the "green" secondary phototrophs, the chlorarachniophytes, suggests multiple horizontal gene transfers from the red lineage before the divergence of extant chlorarachniophytes
- PMID: 24972019
- PMCID: PMC4074131
- DOI: 10.1371/journal.pone.0101158
Phylogenomic analysis of "red" genes from two divergent species of the "green" secondary phototrophs, the chlorarachniophytes, suggests multiple horizontal gene transfers from the red lineage before the divergence of extant chlorarachniophytes
Abstract
The plastids of chlorarachniophytes were derived from an ancestral green alga via secondary endosymbiosis. Thus, genes from the "green" lineage via secondary endosymbiotic gene transfer (EGT) are expected in the nuclear genomes of the Chlorarachniophyta. However, several recent studies have revealed the presence of "red" genes in their nuclear genomes. To elucidate the origin of such "red" genes in chlorarachniophyte nuclear genomes, we carried out exhaustive single-gene phylogenetic analyses, including two operational taxonomic units (OTUs) that represent two divergent sister lineages of the Chlorarachniophyta, Amorphochlora amoeboformis ( = Lotharella amoeboformis; based on RNA sequences newly determined here) and Bigelowiella natans (based on the published genome sequence). We identified 10 genes of cyanobacterial origin, phylogenetic analysis of which showed the chlorarachniophytes to branch with the red lineage (red algae and/or red algal secondary or tertiary plastid-containing eukaryotes). Of the 10 genes, 7 demonstrated robust monophyly of the two chlorarachniophyte OTUs. Thus, the common ancestor of the extant chlorarachniophytes likely experienced multiple horizontal gene transfers from the red lineage. Because 4 of the 10 genes are obviously photosynthesis- and/or plastid-related, and almost all of the eukaryotic OTUs in the 10 trees possess plastids, such red genes most likely originated directly from photosynthetic eukaryotes. This situation could be explained by a possible cryptic endosymbiosis of a red algal plastid before the secondary endosymbiosis of the green algal plastid, or a long-term feeding on a single (or multiple closely related) red algal plastid-containing eukaryote(s) after the green secondary endosymbiosis.
Conflict of interest statement
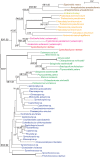
Figures


Similar articles
-
An extended phylogenetic analysis reveals ancient origin of "non-green" phosphoribulokinase genes from two lineages of "green" secondary photosynthetic eukaryotes: Euglenophyta and Chlorarachniophyta.BMC Res Notes. 2011 Sep 7;4:330. doi: 10.1186/1756-0500-4-330. BMC Res Notes. 2011. PMID: 21899749 Free PMC article.
-
The phylogenetic position of red algae revealed by multiple nuclear genes from mitochondria-containing eukaryotes and an alternative hypothesis on the origin of plastids.J Mol Evol. 2003 Apr;56(4):485-97. doi: 10.1007/s00239-002-2419-9. J Mol Evol. 2003. PMID: 12664168
-
A new scenario of plastid evolution: plastid primary endosymbiosis before the divergence of the "Plantae," emended.J Plant Res. 2005 Aug;118(4):247-55. doi: 10.1007/s10265-005-0219-1. Epub 2005 Jul 20. J Plant Res. 2005. PMID: 16032387 Review.
-
Phylogenomic analysis identifies red algal genes of endosymbiotic origin in the chromalveolates.Mol Biol Evol. 2006 Mar;23(3):663-74. doi: 10.1093/molbev/msj075. Epub 2005 Dec 15. Mol Biol Evol. 2006. PMID: 16357039
-
The endosymbiotic origin, diversification and fate of plastids.Philos Trans R Soc Lond B Biol Sci. 2010 Mar 12;365(1541):729-48. doi: 10.1098/rstb.2009.0103. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20124341 Free PMC article. Review.
Cited by
-
Cymbomonas tetramitiformis - a peculiar prasinophyte with a taste for bacteria sheds light on plastid evolution.Symbiosis. 2017;71(1):1-7. doi: 10.1007/s13199-016-0464-1. Epub 2016 Nov 10. Symbiosis. 2017. PMID: 28066124 Free PMC article. Review.
-
Plastid phylogenomics with broad taxon sampling further elucidates the distinct evolutionary origins and timing of secondary green plastids.Sci Rep. 2018 Jan 24;8(1):1523. doi: 10.1038/s41598-017-18805-w. Sci Rep. 2018. PMID: 29367699 Free PMC article.
-
Secondary Plastids of Euglenids and Chlorarachniophytes Function with a Mix of Genes of Red and Green Algal Ancestry.Mol Biol Evol. 2018 Sep 1;35(9):2198-2204. doi: 10.1093/molbev/msy121. Mol Biol Evol. 2018. PMID: 29924337 Free PMC article.
-
Extensive horizontal gene transfer, duplication, and loss of chlorophyll synthesis genes in the algae.BMC Evol Biol. 2015 Feb 10;15:16. doi: 10.1186/s12862-015-0286-4. BMC Evol Biol. 2015. PMID: 25887237 Free PMC article.
-
Evolution of the Tetrapyrrole Biosynthetic Pathway in Secondary Algae: Conservation, Redundancy and Replacement.PLoS One. 2016 Nov 18;11(11):e0166338. doi: 10.1371/journal.pone.0166338. eCollection 2016. PLoS One. 2016. PMID: 27861576 Free PMC article.
References
-
- Reyes-Prieto A, Weber AP, Bhattacharya D (2007) The origin and establishment of the plastid in algae and plants. Annu Rev Genet 41: 147–168. - PubMed
-
- Gould SB, Waller RF, McFadden GI (2008) Plastid evolution. Annu Rev Plant Biol 59: 491–517. - PubMed
-
- Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D (2004) A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol 21: 809–818. - PubMed
-
- Adl SM, Simpson AGB, Farmer MA, Andersen RA, Anderson OR, et al. (2005) The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol 52: 399–451. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

