The strictly aerobic yeast Yarrowia lipolytica tolerates loss of a mitochondrial DNA-packaging protein
- PMID: 24972935
- PMCID: PMC4187616
- DOI: 10.1128/EC.00092-14
The strictly aerobic yeast Yarrowia lipolytica tolerates loss of a mitochondrial DNA-packaging protein
Erratum in
-
Correction for Bakkaiova et al., The strictly aerobic yeast Yarrowia lipolytica tolerates loss of a mitochondrial DNA-packaging protein.Eukaryot Cell. 2015 Jan;14(1):113. doi: 10.1128/EC.00246-14. Eukaryot Cell. 2015. PMID: 25550345 Free PMC article. No abstract available.
Abstract
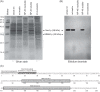
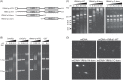
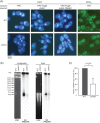
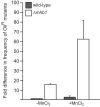
Mitochondrial DNA (mtDNA) is highly compacted into DNA-protein structures termed mitochondrial nucleoids (mt-nucleoids). The key mt-nucleoid components responsible for mtDNA condensation are HMG box-containing proteins such as mammalian mitochondrial transcription factor A (TFAM) and Abf2p of the yeast Saccharomyces cerevisiae. To gain insight into the function and organization of mt-nucleoids in strictly aerobic organisms, we initiated studies of these DNA-protein structures in Yarrowia lipolytica. We identified a principal component of mt-nucleoids in this yeast and termed it YlMhb1p (Y. lipolytica mitochondrial HMG box-containing protein 1). YlMhb1p contains two putative HMG boxes contributing both to DNA binding and to its ability to compact mtDNA in vitro. Phenotypic analysis of a Δmhb1 strain lacking YlMhb1p resulted in three interesting findings. First, although the mutant exhibits clear differences in mt-nucleoids accompanied by a large decrease in the mtDNA copy number and the number of mtDNA-derived transcripts, its respiratory characteristics and growth under most of the conditions tested are indistinguishable from those of the wild-type strain. Second, our results indicate that a potential imbalance between subunits of the respiratory chain encoded separately by nuclear DNA and mtDNA is prevented at a (post)translational level. Third, we found that mtDNA in the Δmhb1 strain is more prone to mutations, indicating that mtHMG box-containing proteins protect the mitochondrial genome against mutagenic events.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Miyakawa I, Aoi H, Sando N, Kuroiwa T. 1984. Fluorescence microscopic studies of mitochondrial nucleoids during meiosis and sporulation in the yeast, Saccharomyces cerevisiae. J. Cell Sci. 66:21–38 - PubMed
-
- Miyakawa I, Sando N, Kawano S, Nakamura S, Kuroiwa T. 1987. Isolation of morphologically intact mitochondrial nucleoids from the yeast, Saccharomyces cerevisiae. J. Cell Sci. 88:431–439 - PubMed
-
- Miyakawa I, Miyamoto M, Kuroiwa T, Sando N. 2004. DNA content of individual mitochondrial nucleoids varies depending on the culture conditions of the yeast Saccharomyces cerevisiae. Cytologia 69:101–107. 10.1508/cytologia.69.101 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

