Synthesis of 53 tissue and cell line expression QTL datasets reveals master eQTLs
- PMID: 24973796
- PMCID: PMC4102726
- DOI: 10.1186/1471-2164-15-532
Synthesis of 53 tissue and cell line expression QTL datasets reveals master eQTLs
Abstract
Background: Gene expression genetic studies in human tissues and cells identify cis- and trans-acting expression quantitative trait loci (eQTLs). These eQTLs provide insights into regulatory mechanisms underlying disease risk. However, few studies systematically characterized eQTL results across cell and tissues types. We synthesized eQTL results from >50 datasets, including new primary data from human brain, peripheral plaque and kidney samples, in order to discover features of human eQTLs.
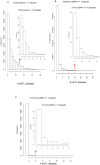
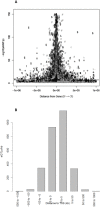
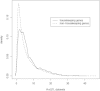
Results: We find a substantial number of robust cis-eQTLs and far fewer trans-eQTLs consistent across tissues. Analysis of 45 full human GWAS scans indicates eQTLs are enriched overall, and above nSNPs, among positive statistical signals in genetic mapping studies, and account for a significant fraction of the strongest human trait effects. Expression QTLs are enriched for gene centricity, higher population allele frequencies, in housekeeping genes, and for coincidence with regulatory features, though there is little evidence of 5' or 3' positional bias. Several regulatory categories are not enriched including microRNAs and their predicted binding sites and long, intergenic non-coding RNAs. Among the most tissue-ubiquitous cis-eQTLs, there is enrichment for genes involved in xenobiotic metabolism and mitochondrial function, suggesting these eQTLs may have adaptive origins. Several strong eQTLs (CDK5RAP2, NBPFs) coincide with regions of reported human lineage selection. The intersection of new kidney and plaque eQTLs with related GWAS suggest possible gene prioritization. For example, butyrophilins are now linked to arterial pathogenesis via multiple genetic and expression studies. Expression QTL and GWAS results are made available as a community resource through the NHLBI GRASP database [http://apps.nhlbi.nih.gov/grasp/].
Conclusions: Expression QTLs inform the interpretation of human trait variability, and may account for a greater fraction of phenotypic variability than protein-coding variants. The synthesis of available tissue eQTL data highlights many strong cis-eQTLs that may have important biologic roles and could serve as positive controls in future studies. Our results indicate some strong tissue-ubiquitous eQTLs may have adaptive origins in humans. Efforts to expand the genetic, splicing and tissue coverage of known eQTLs will provide further insights into human gene regulation.
Figures


Similar articles
-
Conditional entropy in variation-adjusted windows detects selection signatures associated with expression quantitative trait loci (eQTLs).BMC Genomics. 2015;16 Suppl 8(Suppl 8):S8. doi: 10.1186/1471-2164-16-S8-S8. Epub 2015 Jun 18. BMC Genomics. 2015. PMID: 26111110 Free PMC article.
-
The impact of cell type and context-dependent regulatory variants on human immune traits.Genome Biol. 2021 Apr 29;22(1):122. doi: 10.1186/s13059-021-02334-x. Genome Biol. 2021. PMID: 33926512 Free PMC article.
-
Cell-type-specific meQTLs extend melanoma GWAS annotation beyond eQTLs and inform melanocyte gene-regulatory mechanisms.Am J Hum Genet. 2021 Sep 2;108(9):1631-1646. doi: 10.1016/j.ajhg.2021.06.018. Epub 2021 Jul 21. Am J Hum Genet. 2021. PMID: 34293285 Free PMC article.
-
Expression Quantitative Trait Loci Information Improves Predictive Modeling of Disease Relevance of Non-Coding Genetic Variation.PLoS One. 2015 Oct 16;10(10):e0140758. doi: 10.1371/journal.pone.0140758. eCollection 2015. PLoS One. 2015. PMID: 26474488 Free PMC article. Review.
-
Integrated gene expression profiling and linkage analysis in the rat.Mamm Genome. 2006 Jun;17(6):480-9. doi: 10.1007/s00335-005-0181-1. Epub 2006 Jun 12. Mamm Genome. 2006. PMID: 16783629 Review.
Cited by
-
Integromic analysis of genetic variation and gene expression identifies networks for cardiovascular disease phenotypes.Circulation. 2015 Feb 10;131(6):536-49. doi: 10.1161/CIRCULATIONAHA.114.010696. Epub 2014 Dec 22. Circulation. 2015. PMID: 25533967 Free PMC article.
-
Meta-analysis of rare and common exome chip variants identifies S1PR4 and other loci influencing blood cell traits.Nat Genet. 2016 Aug;48(8):867-76. doi: 10.1038/ng.3607. Epub 2016 Jul 11. Nat Genet. 2016. PMID: 27399967 Free PMC article.
-
Genetic Regulation of Adipose Gene Expression and Cardio-Metabolic Traits.Am J Hum Genet. 2017 Mar 2;100(3):428-443. doi: 10.1016/j.ajhg.2017.01.027. Am J Hum Genet. 2017. PMID: 28257690 Free PMC article.
-
Identification of common genetic variants controlling transcript isoform variation in human whole blood.Nat Genet. 2015 Apr;47(4):345-52. doi: 10.1038/ng.3220. Epub 2015 Feb 16. Nat Genet. 2015. PMID: 25685889 Free PMC article.
-
Biological characterization of expression quantitative trait loci (eQTLs) showing tissue-specific opposite directional effects.Eur J Hum Genet. 2019 Nov;27(11):1745-1756. doi: 10.1038/s41431-019-0468-4. Epub 2019 Jul 11. Eur J Hum Genet. 2019. PMID: 31296926 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

