Modulation of age- and cancer-associated DNA methylation change in the healthy colon by aspirin and lifestyle
- PMID: 24973978
- PMCID: PMC4112799
- DOI: 10.1093/jnci/dju161
Modulation of age- and cancer-associated DNA methylation change in the healthy colon by aspirin and lifestyle
Abstract
Background: Aberrant DNA methylation in gene promoters is associated with aging and cancer, but the circumstances determining methylation change are unknown. We investigated the impact of lifestyle modulators of colorectal cancer (CRC) risk on the stability of gene promoter methylation in the colonic mucosa.
Methods: We measured genome-wide promoter CpG methylation in normal colon biopsies (n = 1092) from a female screening cohort, investigated the interaction of lifestyle factors with age-dependent increase in methylation with log-linear multivariable regression, and related their modifying effect to hypermethylation in CRC. All statistical tests were two-sided.
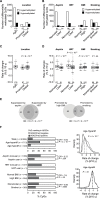
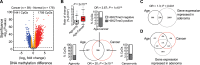
Results: Of 20025 promoter-associated CpGs analyzed, 1713 showed statistically significant age-dependent methylation gains. Fewer CpGs acquired methylation in users of aspirin (≥ 2 years) and hormonal replacement therapy (HRT age ≥ 50 years) compared with nonusers (43 vs 1355; 1 vs1377, respectively), whereas more CpGs were affected in smokers (≥ 20 years) and individuals with a body mass index (BMI) of 25 kg/m(2) and greater compared with control groups (180 vs 39; 554 vs 144, respectively). Fifty percent of the CpGs showing age-dependent methylation were found hypermethylated in CRC (odds ratio [OR] = 20; 95% confidence interval [CI] = 18 to 23; P < 2 × 10(-16)). These loci gained methylation with a higher median rate compared with age-only methylated sites (P = 2 × 10(-76)) and were enriched for polycomb regions (OR = 3.67). Importantly, aspirin (P < .001) and HRT use (P < .001) reduced the methylation rate at these cancer-related genes, whereas smoking (P < .001) and high BMI (P = .004) increased it.
Conclusions: Lifestyle, including aspirin use, modulates age-associated DNA methylation change in the colonic epithelium and thereby impacts the evolution of cancer methylomes.
© The Author 2014. Published by Oxford University Press.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

