Transcriptome dynamics of Arabidopsis thaliana root penetration by the oomycete pathogen Phytophthora parasitica
- PMID: 24974100
- PMCID: PMC4111850
- DOI: 10.1186/1471-2164-15-538
Transcriptome dynamics of Arabidopsis thaliana root penetration by the oomycete pathogen Phytophthora parasitica
Abstract
Background: Oomycetes are a group of filamentous microorganisms that includes both animal and plant pathogens and causes major agricultural losses. Phytophthora species can infect most crops and plants from natural ecosystems. Despite their tremendous economic and ecologic importance, few effective methods exist for limiting the damage caused by these species. New solutions are required, and their development will require improvements in our understanding of the molecular events governing infection by these pathogens. In this study, we characterized the genetic program activated during penetration of the plant by the soil-borne pathogen Phytophthora parasitica.
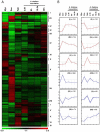
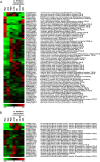
Results: Using all the P. parasitica sequences available in public databases, we generated a custom oligo-array and performed a transcriptomic analysis of the early events of Arabidopsis thaliana infection. We characterized biological stages, ranging from the appressorium-mediated penetration of the pathogen into the roots to the occurrence of first dead cells in the plant. We identified a series of sequences that were transiently modulated during host penetration. Surprisingly, we observed an overall down regulation of genes encoding proteins involved in lipid and sugar metabolism, and an upregulation of functions controlling the transport of amino acids. We also showed that different groups of genes were expressed by P. parasitica during host penetration and the subsequent necrotrophic phase. Differential expression patterns were particularly marked for cell wall-degrading enzymes and other proteins involved in pathogenicity, including RXLR effectors. By transforming P. parasitica with a transcriptional fusion with GFP, we showed that an RXLR-ecoding gene was expressed in the appressorium and infectious hyphae during infection of the first plant cell.
Conclusion: We have characterized the genetic program activated during the initial invasion of plant cells by P. parasitica. We showed that a specific set of proteins, including effectors, was mobilized for penetration and to facilitate infection. Our detection of the expression of an RXLR encoding gene by the appressorium and infection hyphae highlights a role of this structure in the manipulation of the host cells.
Figures


References
-
- Erwin DC, Ribeiro OK. Phytophthora Diseases Worldwide. St. Paul, MN: American Phytopathological Society; 1996.
-
- Swiecki TJ, Donald M. Histology of chrysanthemum roots exposed to salinity stress and Phytophthora cryptogea. Can J Bot. 1988;66:280–288. doi: 10.1139/b88-046. - DOI
-
- Dale ML, Irwin JAG. Stomata as an infection court for Phytophtora megasperma f. sp. medicaginis in chickpea and a histological study of infection. Phytopathology. 1991;91:375–379. doi: 10.1094/Phyto-81-375. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

