Biosynthesis of polybrominated aromatic organic compounds by marine bacteria
- PMID: 24974229
- PMCID: PMC4104138
- DOI: 10.1038/nchembio.1564
Biosynthesis of polybrominated aromatic organic compounds by marine bacteria
Abstract
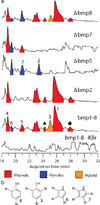
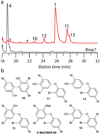
Polybrominated diphenyl ethers (PBDEs) and polybrominated bipyrroles are natural products that bioaccumulate in the marine food chain. PBDEs have attracted widespread attention because of their persistence in the environment and potential toxicity to humans. However, the natural origins of PBDE biosynthesis are not known. Here we report marine bacteria as producers of PBDEs and establish a genetic and molecular foundation for their production that unifies paradigms for the elaboration of bromophenols and bromopyrroles abundant in marine biota. We provide biochemical evidence of marine brominases revealing decarboxylative-halogenation enzymology previously unknown among halogenating enzymes. Biosynthetic motifs discovered in our study were used to mine sequence databases to discover unrealized marine bacterial producers of organobromine compounds.
Figures




References
-
- Gribble GW. The natural production of organobromine compounds. Environ Sci Pollut Res Int. 2000;7:37–47. - PubMed
-
- Gribble GW. Naturally Occurring Organohalogen Compounds - A Comprehensive Update. Vol. 91. Springer Vienna; 2010.
-
- Liu YN, et al. Spatial and temporal distributions of bromoform and dibromomethane in the Atlantic Ocean and their relationship with photosynthetic biomass. J Geophys Res-Oceans. 2013;118:3950–3965.
-
- Gaul S, et al. Identification of the natural product 2,3,4,5-tetrabromo-1-methylpyrrole in Pacific biota, passive samplers and seagrass from Queensland, Australia. Mar Pollut Bull. 2011;62:2463–2468. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- PubChem-Substance/182111901
- PubChem-Substance/182111902
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

