Global analysis of cellular proteolysis by selective enzymatic labeling of protein N-termini
- PMID: 24974296
- PMCID: PMC4260929
- DOI: 10.1016/B978-0-12-417158-9.00013-3
Global analysis of cellular proteolysis by selective enzymatic labeling of protein N-termini
Abstract
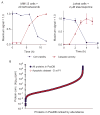
Proteolysis is a critical modification leading to alteration of protein function with important outcomes in many biological processes. However, for the majority of proteases, we have an incomplete understanding of both cellular substrates and downstream effects. Here, we describe detailed protocols and applications for using the rationally engineered peptide ligase, subtiligase, to specifically label and capture protein N-termini generated by proteases either induced or added to complex biological samples. This method allows identification of the protein targets as well as their precise cleavage locations. This approach has revealed >8000 proteolytic sites in healthy and apoptotic cells including >1700 caspase cleavages. One can further determine substrate preferences through rate analysis with quantitative mass spectrometry, physiological substrate specificities, and even infer the identity of proteases operating in the cell. In this chapter, we also describe how this experimental method can be generalized to investigate proteolysis in any biological sample.
Keywords: Apoptosis; Caspase; Degradomics; Mass spectrometry; Proteolysis; Proteomics; Selected reaction monitoring; Subtiligase.
© 2014 Elsevier Inc. All rights reserved.
Figures

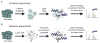




Similar articles
-
N-Terminal Modification of Proteins with Subtiligase Specificity Variants.Curr Protoc Chem Biol. 2020 Mar;12(1):e79. doi: 10.1002/cpch.79. Curr Protoc Chem Biol. 2020. PMID: 32074409
-
The DegraBase: a database of proteolysis in healthy and apoptotic human cells.Mol Cell Proteomics. 2013 Mar;12(3):813-24. doi: 10.1074/mcp.O112.024372. Epub 2012 Dec 20. Mol Cell Proteomics. 2013. PMID: 23264352 Free PMC article.
-
Comparative assessment of large-scale proteomic studies of apoptotic proteolysis.ACS Chem Biol. 2009 Jun 19;4(6):401-8. doi: 10.1021/cb900082q. ACS Chem Biol. 2009. PMID: 19415908 Free PMC article.
-
Protein TAILS: when termini tell tales of proteolysis and function.Curr Opin Chem Biol. 2013 Feb;17(1):73-82. doi: 10.1016/j.cbpa.2012.11.025. Epub 2013 Jan 6. Curr Opin Chem Biol. 2013. PMID: 23298954 Review.
-
MS-driven protease substrate degradomics.Proteomics. 2010 Mar;10(6):1284-96. doi: 10.1002/pmic.200900418. Proteomics. 2010. PMID: 20058249 Review.
Cited by
-
N-Terminomics Strategies for Protease Substrates Profiling.Molecules. 2021 Aug 3;26(15):4699. doi: 10.3390/molecules26154699. Molecules. 2021. PMID: 34361849 Free PMC article. Review.
-
Engineering peptide ligase specificity by proteomic identification of ligation sites.Nat Chem Biol. 2018 Jan;14(1):50-57. doi: 10.1038/nchembio.2521. Epub 2017 Nov 20. Nat Chem Biol. 2018. PMID: 29155430 Free PMC article.
-
Deep profiling of protease substrate specificity enabled by dual random and scanned human proteome substrate phage libraries.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25464-25475. doi: 10.1073/pnas.2009279117. Epub 2020 Sep 24. Proc Natl Acad Sci U S A. 2020. PMID: 32973096 Free PMC article.
-
An optimized quantitative proteomics method establishes the cell type-resolved mouse brain secretome.EMBO J. 2020 Oct 15;39(20):e105693. doi: 10.15252/embj.2020105693. Epub 2020 Sep 21. EMBO J. 2020. PMID: 32954517 Free PMC article.
-
Mass spectrometry-based detection and assignment of protein posttranslational modifications.ACS Chem Biol. 2015 Jan 16;10(1):63-71. doi: 10.1021/cb500904b. ACS Chem Biol. 2015. PMID: 25541750 Free PMC article. Review.
References
-
- Abrahmsen L, Tom J, Burnier J, Butcher KA, Kossiakoff A, Wells JA. Engineering subtilisin and its substrates for efficient ligation of peptide bonds in aqueous solution. Biochemistry. 1991;30:4151–4159. - PubMed
-
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

