Identification of cardiac myofilament protein isoforms using multiple mass spectrometry based approaches
- PMID: 24974818
- PMCID: PMC4222538
- DOI: 10.1002/prca.201400039
Identification of cardiac myofilament protein isoforms using multiple mass spectrometry based approaches
Abstract
Purpose: The identification of protein isoforms in complex biological samples is challenging. We, therefore, used an MS approach to unambiguously identify cardiac myofilament protein isoforms based on the observation of a tryptic peptide consisting of a sequence unique to a particular isoform.
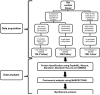
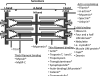
Experimental design: Three different workflows were used to isolate and fractionate rat cardiac myofilament subproteomes. All fractions were analyzed on an LTQ-Orbitrap MS, proteins were identified using various search engines (MASCOT, X!Tandem, X!Tandem Kscore, and OMSSA) with results combined via PepArML Meta-Search engine, and a postsearch analysis was performed by MASPECTRAS. All MS data have been deposited in the ProteomeXchange with identifier PXD000874 (http://proteomecentral.proteomexchange.org/dataset/PXD000874).
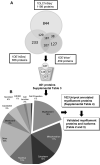
Results: The combination of multiple workflows and search engines resulted in a larger number of nonredundant proteins identified than with individual methods. A total of 102 myofilament annotated proteins were observed overlapping in two or three of the workflows. Literature search for myofilament presence with manual validation of the MS spectra was carried out for unambiguous identification: ten cardiac myofilament and 17 cardiac myofilament-associated proteins were identified with 39 isoforms and subisoforms.
Conclusion and clinical relevance: We have identified multiple isoforms of myofilament proteins that are present in cardiac tissue using unique tryptic peptides. Changes in distribution of these protein isoforms under pathological conditions could ultimately allow for clinical diagnostics or as therapeutic targets.
Keywords: Cardiac; Myofilament proteins; Protein isoforms; Protein subisoforms.
© 2014 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures


Similar articles
-
In-depth analysis of protein inference algorithms using multiple search engines and well-defined metrics.J Proteomics. 2017 Jan 6;150:170-182. doi: 10.1016/j.jprot.2016.08.002. Epub 2016 Aug 4. J Proteomics. 2017. PMID: 27498275
-
Top-down mass spectrometry of cardiac myofilament proteins in health and disease.Proteomics Clin Appl. 2014 Aug;8(7-8):554-68. doi: 10.1002/prca.201400043. Proteomics Clin Appl. 2014. PMID: 24945106 Free PMC article. Review.
-
Top-down proteomics reveals concerted reductions in myofilament and Z-disc protein phosphorylation after acute myocardial infarction.Mol Cell Proteomics. 2014 Oct;13(10):2752-64. doi: 10.1074/mcp.M114.040675. Epub 2014 Jun 26. Mol Cell Proteomics. 2014. PMID: 24969035 Free PMC article.
-
Top-Down Proteomics Reveals Myofilament Proteoform Heterogeneity among Various Rat Skeletal Muscle Tissues.J Proteome Res. 2020 Jan 3;19(1):446-454. doi: 10.1021/acs.jproteome.9b00623. Epub 2019 Nov 7. J Proteome Res. 2020. PMID: 31647247 Free PMC article.
-
Targeted proteomics of myofilament phosphorylation and other protein posttranslational modifications.Proteomics Clin Appl. 2014 Aug;8(7-8):543-53. doi: 10.1002/prca.201400034. Proteomics Clin Appl. 2014. PMID: 24976615 Review.
Cited by
-
Myofilament-associated proteins with intrinsic disorder (MAPIDs) and their resolution by computational modeling.Q Rev Biophys. 2023 Jan 11;56:e2. doi: 10.1017/S003358352300001X. Q Rev Biophys. 2023. PMID: 36628457 Free PMC article. Review.
-
AQUA Mutant Protein Quantification of Endomyocardial Biopsy-Sized Samples From a Patient With Hypertrophic Cardiomyopathy.Front Cardiovasc Med. 2022 Feb 21;9:816330. doi: 10.3389/fcvm.2022.816330. eCollection 2022. Front Cardiovasc Med. 2022. PMID: 35265683 Free PMC article.
-
Proteomics of the heart.Physiol Rev. 2024 Jul 1;104(3):931-982. doi: 10.1152/physrev.00026.2023. Epub 2024 Feb 1. Physiol Rev. 2024. PMID: 38300522 Free PMC article. Review.
-
Myofilament Phosphorylation in Stem Cell Treated Diastolic Heart Failure.Circ Res. 2021 Dec 3;129(12):1125-1140. doi: 10.1161/CIRCRESAHA.119.316311. Epub 2021 Oct 13. Circ Res. 2021. PMID: 34641704 Free PMC article.
-
Conservation and divergence of protein pathways in the vertebrate heart.PLoS Biol. 2019 Sep 6;17(9):e3000437. doi: 10.1371/journal.pbio.3000437. eCollection 2019 Sep. PLoS Biol. 2019. PMID: 31490923 Free PMC article.
References
-
- Gunning PW. Protein Isoforms and Isozymes. eLS. 2006
-
- Marston SB, Redwood CS. Modulation of thin filament activation by breakdown or isoform switching of thin filament proteins: physiological and pathological implications. Circ.Res. 2003;93:1170–1178. - PubMed
-
- Wolska BM, Wieczorek DM. The role of tropomyosin in the regulation of myocardial contraction and relaxation. Pflugers Arch. 2003;446:1–8. - PubMed
-
- Denz CR, Narshi A, Zajdel RW, Dube DK. Expression of a novel cardiac-specific tropomyosin isoform in humans. Biochem.Biophys.Res.Commun. 2004;320:1291–1297. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

