Docking server for the identification of heparin binding sites on proteins
- PMID: 24974889
- PMCID: PMC4184157
- DOI: 10.1021/ci500115j
Docking server for the identification of heparin binding sites on proteins
Abstract
Many proteins of widely differing functionality and structure are capable of binding heparin and heparan sulfate. Since crystallizing protein-heparin complexes for structure determination is generally difficult, computational docking can be a useful approach for understanding specific interactions. Previous studies used programs originally developed for docking small molecules to well-defined pockets, rather than for docking polysaccharides to highly charged shallow crevices that usually bind heparin. We have extended the program PIPER and the automated protein-protein docking server ClusPro to heparin docking. Using a molecular mechanics energy function for scoring and the fast Fourier transform correlation approach, the method generates and evaluates close to a billion poses of a heparin tetrasaccharide probe. The docked structures are clustered using pairwise root-mean-square deviations as the distance measure. It was shown that clustering of heparin molecules close to each other but having different orientations and selecting the clusters with the highest protein-ligand contacts reliably predicts the heparin binding site. In addition, the centers of the five most populated clusters include structures close to the native orientation of the heparin. These structures can provide starting points for further refinement by methods that account for flexibility such as molecular dynamics. The heparin docking method is available as an advanced option of the ClusPro server at http://cluspro.bu.edu/ .
Figures

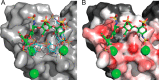
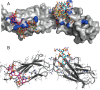
Similar articles
-
The ClusPro web server for protein-protein docking.Nat Protoc. 2017 Feb;12(2):255-278. doi: 10.1038/nprot.2016.169. Epub 2017 Jan 12. Nat Protoc. 2017. PMID: 28079879 Free PMC article.
-
Achieving reliability and high accuracy in automated protein docking: ClusPro, PIPER, SDU, and stability analysis in CAPRI rounds 13-19.Proteins. 2010 Nov 15;78(15):3124-30. doi: 10.1002/prot.22835. Proteins. 2010. PMID: 20818657 Free PMC article.
-
New additions to the ClusPro server motivated by CAPRI.Proteins. 2017 Mar;85(3):435-444. doi: 10.1002/prot.25219. Epub 2017 Jan 5. Proteins. 2017. PMID: 27936493 Free PMC article.
-
Interactions of heparin/heparan sulfate with proteins: appraisal of structural factors and experimental approaches.Glycobiology. 2004 Apr;14(4):17R-30R. doi: 10.1093/glycob/cwh051. Epub 2004 Jan 12. Glycobiology. 2004. PMID: 14718374 Review.
-
Heparin-protein interactions.Angew Chem Int Ed Engl. 2002 Feb 1;41(3):391-412. doi: 10.1002/1521-3773(20020201)41:3<390::aid-anie390>3.0.co;2-b. Angew Chem Int Ed Engl. 2002. PMID: 12491369 Review.
Cited by
-
Convergent chemoenzymatic synthesis and biological evaluation of a heparan sulfate proteoglycan syndecan-1 mimetic.Chem Commun (Camb). 2021 Apr 7;57(27):3407-3410. doi: 10.1039/d1cc00796c. Epub 2021 Mar 9. Chem Commun (Camb). 2021. PMID: 33687395 Free PMC article.
-
Estimating glycosaminoglycan-protein interaction affinity: water dominates the specific antithrombin-heparin interaction.Glycobiology. 2016 Oct;26(10):1041-1047. doi: 10.1093/glycob/cww073. Epub 2016 Jul 18. Glycobiology. 2016. PMID: 27496757 Free PMC article.
-
Heparin-Induced Changes of Vascular Endothelial Growth Factor (VEGF165) Structure.Biomolecules. 2023 Jan 3;13(1):98. doi: 10.3390/biom13010098. Biomolecules. 2023. PMID: 36671483 Free PMC article.
-
Three-Dimensional Structures of Carbohydrates and Where to Find Them.Int J Mol Sci. 2020 Oct 18;21(20):7702. doi: 10.3390/ijms21207702. Int J Mol Sci. 2020. PMID: 33081008 Free PMC article. Review.
-
The ClusPro web server for protein-protein docking.Nat Protoc. 2017 Feb;12(2):255-278. doi: 10.1038/nprot.2016.169. Epub 2017 Jan 12. Nat Protoc. 2017. PMID: 28079879 Free PMC article.
References
-
- Bernfield M.; Gotte M.; Park P. W.; Reizes O.; Fitzgerald M. L.; Lincecum J.; Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 1999, 68, 729–77. - PubMed
-
- Gandhi N. S.; Mancera R. L. The structure of glycosaminoglycans and their interactions with proteins. Chem. Biol. Drug Des 2008, 72, 455–482. - PubMed
-
- Lindahl U. Heparan sulfate-protein interactions - a concept for drug design?. Thromb Haemostasis 2007, 98, 109–115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

