Transcriptomic and physiological characterization of the fefe mutant of melon (Cucumis melo) reveals new aspects of iron-copper crosstalk
- PMID: 24975482
- PMCID: PMC4117724
- DOI: 10.1111/nph.12911
Transcriptomic and physiological characterization of the fefe mutant of melon (Cucumis melo) reveals new aspects of iron-copper crosstalk
Abstract
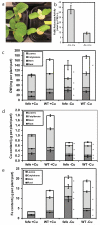
Iron (Fe) and copper (Cu) homeostasis are tightly linked across biology. In previous work, Fe deficiency interacted with Cu-regulated genes and stimulated Cu accumulation. The C940-fe (fefe) Fe-uptake mutant of melon (Cucumis melo) was characterized, and the fefe mutant was used to test whether Cu deficiency could stimulate Fe uptake. Wild-type and fefe mutant transcriptomes were determined by RNA-seq under Fe and Cu deficiency. FeFe-regulated genes included core Fe uptake, metal homeostasis, and transcription factor genes. Numerous genes were regulated by both Fe and Cu. The fefe mutant was rescued by high Fe or by Cu deficiency, which stimulated ferric-chelate reductase activity, FRO2 expression, and Fe accumulation. Accumulation of Fe in Cu-deficient plants was independent of the normal Fe-uptake system. One of the four FRO genes in the melon and cucumber (Cucumis sativus) genomes was Fe-regulated, and one was Cu-regulated. Simultaneous Fe and Cu deficiency synergistically up-regulated Fe-uptake gene expression. Overlap in Fe and Cu deficiency transcriptomes highlights the importance of Fe-Cu crosstalk in metal homeostasis. The fefe gene is not orthologous to FIT, and thus identification of this gene will provide clues to help understand regulation of Fe uptake in plants.
Keywords: copper (Cu); fefe mutant; ferric-chelate reductase; iron (Fe); iron-copper crosstalk; melon (Cucumis melo); metal homeostasis.
© 2014 The Authors. New Phytologist © 2014 New Phytologist Trust.
Figures









References
-
- Alscher RG, Erturk N, Heath LS. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. Journal of Experimental Botany. 2002;53:1331–1341. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300.
-
- Bernal M, Casero D, Singh V, Wilson GT, Grande A, Yang H, Dodani SC, Pellegrini M, Huijser P, Connolly EL, et al. Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis. The Plant Cell. 2012;24:738–761. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

