An adaptive permutation approach for genome-wide association study: evaluation and recommendations for use
- PMID: 24976866
- PMCID: PMC4070098
- DOI: 10.1186/1756-0381-7-9
An adaptive permutation approach for genome-wide association study: evaluation and recommendations for use
Abstract
Background: Permutation testing is a robust and popular approach for significance testing in genomic research, which has the broad advantage of estimating significance non-parametrically, thereby safe guarding against inflated type I error rates. However, the computational efficiency remains a challenging issue that limits its wide application, particularly in genome-wide association studies (GWAS). Because of this, adaptive permutation strategies can be employed to make permutation approaches feasible. While these approaches have been used in practice, there is little research into the statistical properties of these approaches, and little guidance into the proper application of such a strategy for accurate p-value estimation at the GWAS level.
Methods: In this work, we advocate an adaptive permutation procedure that is statistically valid as well as computationally feasible in GWAS. We perform extensive simulation experiments to evaluate the robustness of the approach to violations of modeling assumptions and compare the power of the adaptive approach versus standard approaches. We also evaluate the parameter choices in implementing the adaptive permutation approach to provide guidance on proper implementation in real studies. Additionally, we provide an example of the application of adaptive permutation testing on real data.
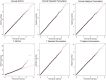
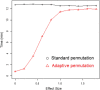
Results: The results provide sufficient evidence that the adaptive test is robust to violations of modeling assumptions. In addition, even when modeling assumptions are correct, the power achieved by adaptive permutation is identical to the parametric approach over a range of significance thresholds and effect sizes under the alternative. A framework for proper implementation of the adaptive procedure is also generated.
Conclusions: While the adaptive permutation approach presented here is not novel, the current study provides evidence of the validity of the approach, and importantly provides guidance on the proper implementation of such a strategy. Additionally, tools are made available to aid investigators in implementing these approaches.
Figures

References
-
- Besag J, Clifford P. Sequential Monte-Carlo p-values. Biometrika. 1991;78(2):301–304. doi: 10.1093/biomet/78.2.301. - DOI
-
- Boos DD, Zhang J. Monte Carlo evaluation of resampling-based hypothesis tests. J Am Stat Assoc. 2000;95(450):486–492. doi: 10.1080/01621459.2000.10474226. - DOI
-
- Phipson B, Smyth GK. Permutation p-values should never be zero: calculating exact p-values when permutations are randomly drawn. Stat Appl Genet Mol Biol. 2010;9:Article39. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

