Association between RUNX3 promoter methylation and non-small cell lung cancer: a meta-analysis
- PMID: 24976992
- PMCID: PMC4073407
- DOI: 10.3978/j.issn.2072-1439.2014.04.09
Association between RUNX3 promoter methylation and non-small cell lung cancer: a meta-analysis
Abstract
Background: Runt-related transcription factor 3 (RUNX3) is a known regulator in the transforming growth factor (TGF)-β signaling pathway, which promoter methylation playing a crucial role in diverse neoplasias. However, the relationship between RUNX3 promoter methylation and non-small cell lung cancer (NSCLC) remains to be clarified.
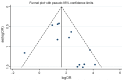
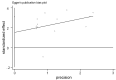
Methods: We searched Pubmed, Embase, Cochrane Central, and Chinese Biological Medicine database, for articles published in English or Chinese until March 7, 2014. Our main analyses were focused on the association between RUNX3 promoter methylation and risk of NSCLC by meta-analysis methods. If heterogeneity was observed, we used random effects model to calculate the overall odds ratios, otherwise fixed effects model was used. Subgroup analyses and meta-regression analyses were employed to detect the sources of the heterogeneity. Sensitivity analysis was performed to evaluate the stability of our studies. A funnel plot and Egger's test were conducted to investigate any potential publication bias.
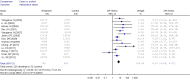
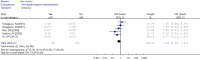
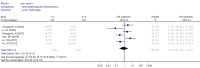
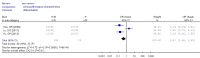
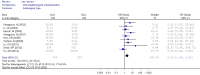
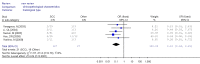
Results: A total of 1,368 samples from 13 literatures were involved in this meta-analysis. The pooled odds ratio (OR) of RUNX3 methylation in NSCLC specimens compared to non-cancer controls was 6.70 [95% confidence interval (CI): 4.64-9.67]. In the analysis of specimen-types subgroup, the summary OR was 5.79 (95% CI: 3.97-8.46) for tissue specimen subgroup, and that was 45.64 (95% CI: 5.89-353.72) for serum specimen subgroup. The ORs for the age ≤60 years, 60-65 years and >65 years subgroup were 5.19 (95% CI: 3.27-8.24), 9.45 (95% CI: 2.45-36.45) and 13.23 (95% CI: 5.59-31.28) respectively. The result of meta-regression indicated that age was fundamental source of heterogeneity (coefficient =0.61, P=0.046, adjusted R(2) =100%). No publication bias was detected. In cancer specimens, the RUNX3 methylation was associated with histological type of the NSCLC, but no significant differences were found for RUNX3 methylation in relation to gender, smoking history, tumor TNM stage or tumor differentiation level.
Conclusions: This meta-analysis of pooled data provides additional evidence to support a strong association between methylation of the RUNX3 promoter and NSCLC. RUNX3 methylation was increasing with age.
Keywords: Non-small cell lung cancer (NSCLC); meta-analysis; methylation; promoter; runt-related transcription factor 3 (RUNX3).
Figures




References
-
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin 2009;59:225-49 - PubMed
-
- Stinchcombe TE, Socinski MA. Current treatments for advanced stage non-small cell lung cancer. Proc Am Thorac Soc 2009;6:233-41 - PubMed
-
- Esteller M, Corn PG, Baylin SB, et al. A gene hypermethylation profile of human cancer. Cancer Res 2001;61:3225-9 - PubMed
-
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002;3:415-28 - PubMed
-
- Risch A, Plass C.Lung cancer epigenetics and genetics. Int J Cancer 2008;123:1-7 - PubMed
LinkOut - more resources
Full Text Sources
