Screening for genes coding for putative antitumor compounds, antimicrobial and enzymatic activities from haloalkalitolerant and haloalkaliphilic bacteria strains of Algerian Sahara Soils
- PMID: 24977147
- PMCID: PMC4058248
- DOI: 10.1155/2014/317524
Screening for genes coding for putative antitumor compounds, antimicrobial and enzymatic activities from haloalkalitolerant and haloalkaliphilic bacteria strains of Algerian Sahara Soils
Abstract
Extreme environments may often contain unusual bacterial groups whose physiology is distinct from those of normal environments. To satisfy the need for new bioactive pharmaceuticals compounds and enzymes, we report here the isolation of novel bacteria from an extreme environment. Thirteen selected haloalkalitolerant and haloalkaliphilic bacteria were isolated from Algerian Sahara Desert soils. These isolates were screened for the presence of genes coding for putative antitumor compounds using PCR based methods. Enzymatic, antibacterial, and antifungal activities were determined by using cultural dependant methods. Several of these isolates are typical of desert and alkaline saline soils, but, in addition, we report for the first time the presence of a potential new member of the genus Nocardia with particular activity against the yeast Saccharomyces cerevisiae. In addition to their haloalkali character, the presence of genes coding for putative antitumor compounds, combined with the antimicrobial activity against a broad range of indicator strains and their enzymatic potential, makes them suitable for biotechnology applications.
Figures



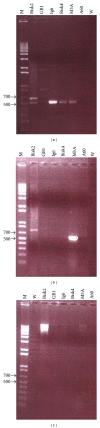

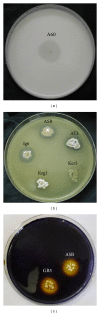
References
-
- Podar M, Reysenbach AL. New opportunities revealed by biotechnological explorations of extremophiles. Current Opinion in Biotechnology. 2006;17(3):250–255. - PubMed
-
- Subramaniam S. Productivity and attrition: key challenges for biotech and pharma. Drug Discovery Today. 2003;8(12):513–515. - PubMed
-
- Lam KS. New aspects of natural products in drug discovery. Trends in Microbiology. 2007;15(6):279–289. - PubMed
-
- Perić-Concha N, Long PF. Mining the microbial metabolome: a new frontier for natural product lead discovery. Drug Discovery Today. 2003;8(23):1078–1084. - PubMed
-
- Spížek J, Novotná J, Řezanka T, Demain AL. Do we need new antibiotics? The search for new targets and new compounds. Journal of Industrial Microbiology and Biotechnology. 2010;37(12):1241–1248. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

