Pathway reconstruction of airway remodeling in chronic lung diseases: a systems biology approach
- PMID: 24978043
- PMCID: PMC4076832
- DOI: 10.1371/journal.pone.0100094
Pathway reconstruction of airway remodeling in chronic lung diseases: a systems biology approach
Abstract
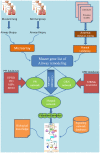
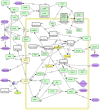
Airway remodeling is a pathophysiologic process at the clinical, cellular, and molecular level relating to chronic obstructive airway diseases such as chronic obstructive pulmonary disease (COPD), asthma and mustard lung. These diseases are associated with the dysregulation of multiple molecular pathways in the airway cells. Little progress has so far been made in discovering the molecular causes of complex disease in a holistic systems manner. Therefore, pathway and network reconstruction is an essential part of a systems biology approach to solve this challenging problem. In this paper, multiple data sources were used to construct the molecular process of airway remodeling pathway in mustard lung as a model of airway disease. We first compiled a master list of genes that change with airway remodeling in the mustard lung disease and then reconstructed the pathway by generating and merging the protein-protein interaction and the gene regulatory networks. Experimental observations and literature mining were used to identify and validate the master list. The outcome of this paper can provide valuable information about closely related chronic obstructive airway diseases which are of great importance for biologists and their future research. Reconstructing the airway remodeling interactome provides a starting point and reference for the future experimental study of mustard lung, and further analysis and development of these maps will be critical to understanding airway diseases in patients.
Conflict of interest statement
Figures


Similar articles
-
The footprint of TGF-β in airway remodeling of the mustard lung.Inhal Toxicol. 2015;27(14):745-53. doi: 10.3109/08958378.2015.1116645. Epub 2015 Nov 26. Inhal Toxicol. 2015. PMID: 26606948 Review.
-
Progress in the development of kinase inhibitors for treating asthma and COPD.Adv Pharmacol. 2023;98:145-178. doi: 10.1016/bs.apha.2023.04.004. Epub 2023 May 3. Adv Pharmacol. 2023. PMID: 37524486 Review.
-
Remodeling in asthma and chronic obstructive pulmonary disease.Proc Am Thorac Soc. 2006 Jul;3(5):434-9. doi: 10.1513/pats.200601-006AW. Proc Am Thorac Soc. 2006. PMID: 16799088 Review.
-
Posttranscriptional Gene Regulatory Networks in Chronic Airway Inflammatory Diseases: In silico Mapping of RNA-Binding Protein Expression in Airway Epithelium.Front Immunol. 2020 Oct 16;11:579889. doi: 10.3389/fimmu.2020.579889. eCollection 2020. Front Immunol. 2020. PMID: 33178205 Free PMC article.
-
Relationship between lung function and quantitative computed tomographic parameters of airway remodeling, air trapping, and emphysema in patients with asthma and chronic obstructive pulmonary disease: A single-center study.J Allergy Clin Immunol. 2016 May;137(5):1413-1422.e12. doi: 10.1016/j.jaci.2016.02.001. Epub 2016 Mar 19. J Allergy Clin Immunol. 2016. PMID: 27006248 Free PMC article.
Cited by
-
Targeting Trefoil Factor Family 3 in Obstructive Airway Diseases: A Computational Approach to Novel Therapeutics.Iran J Med Sci. 2025 Mar 1;50(3):159-170. doi: 10.30476/ijms.2024.101737.3435. eCollection 2025 Mar. Iran J Med Sci. 2025. PMID: 40224201 Free PMC article.
-
Long non-coding RNA TUG1 accelerates abnormal growth of airway smooth muscle cells in asthma by targeting the miR-138-5p/E2F3 axis.Exp Ther Med. 2021 Nov;22(5):1229. doi: 10.3892/etm.2021.10663. Epub 2021 Aug 27. Exp Ther Med. 2021. PMID: 34539825 Free PMC article.
-
Inhibition of SERPINE2/protease nexin-1 by a monoclonal antibody attenuates airway remodeling in a murine model of asthma.Int J Clin Exp Pathol. 2017 Dec 1;10(12):11838-11848. eCollection 2017. Int J Clin Exp Pathol. 2017. PMID: 31966548 Free PMC article.
-
Systems medicine: evolution of systems biology from bench to bedside.Wiley Interdiscip Rev Syst Biol Med. 2015 Jul-Aug;7(4):141-61. doi: 10.1002/wsbm.1297. Epub 2015 Apr 17. Wiley Interdiscip Rev Syst Biol Med. 2015. PMID: 25891169 Free PMC article. Review.
References
-
- Tschumperlin DJ, Drazen JM (2001) Mechanical stimuli to airway remodeling. American journal of respiratory and critical care medicine 164: S90–S94. - PubMed
-
- Thomason JW, Rice TW, Milstone AP (2003) Bronchiolitis obliterans in a survivor of a chemical weapons attack. JAMA 290: 598–599. - PubMed
-
- Ghanei M, Mokhtari M, Mohammad MM, Aslani J (2004) Bronchiolitis obliterans following exposure to sulfur mustard: chest high resolution computed tomography. Eur J Radiol 52: 164–169. - PubMed
-
- Ghanei M, Harandi AA (2007) Long term consequences from exposure to sulfur mustard: a review. Inhal Toxicol 19: 451–456. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

