Differentially expressed plasma microRNAs and the potential regulatory function of Let-7b in chronic thromboembolic pulmonary hypertension
- PMID: 24978044
- PMCID: PMC4076206
- DOI: 10.1371/journal.pone.0101055
Differentially expressed plasma microRNAs and the potential regulatory function of Let-7b in chronic thromboembolic pulmonary hypertension
Abstract
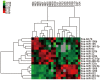
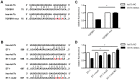
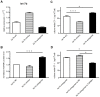
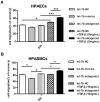
Chronic thromboembolic pulmonary hypertension (CTEPH) is a progressive disease characterized by misguided thrombolysis and remodeling of pulmonary arteries. MicroRNAs are small non-coding RNAs involved in multiple cell processes and functions. During CTEPH, circulating microRNA profile endued with characteristics of diseased cells could be identified as a biomarker, and might help in recognition of pathogenesis. Thus, in this study, we compared the differentially expressed microRNAs in plasma of CTEPH patients and healthy controls and investigated their potential functions. Microarray was used to identify microRNA expression profile and qRT-PCR for validation. The targets of differentially expressed microRNAs were identified in silico, and the Gene Ontology database and Kyoto Encyclopedia of Genes and Genomes pathway database were used for functional investigation of target gene profile. Targets of let-7b were validated by fluorescence reporter assay. Protein expression of target genes was determined by ELISA or western blotting. Cell migration was evaluated by wound healing assay. The results showed that 1) thirty five microRNAs were differentially expressed in CTEPH patients, among which, a signature of 17 microRNAs, which was shown to be related to the disease pathogenesis by in silico analysis, gave diagnostic efficacy of both sensitivity and specificity >0.9. 2) Let-7b, one of the down-regulated anti-oncogenic microRNAs in the signature, was validated to decrease to about 0.25 fold in CTEPH patients. 3) ET-1 and TGFBR1 were direct targets of let-7b. Altering let-7b level influenced ET-1 and TGFBR1 expression in pulmonary arterial endothelial cells (PAECs) as well as the migration of PAECs and pulmonary arterial smooth muscle cells (PASMCs). These results suggested that CTEPH patients had aberrant microRNA signature which might provide some clue for pathogenesis study and biomarker screening. Reduced let-7b might be involved in the pathogenesis of CTEPH by affecting ET-1 expression and the function of PAECs and PASMCs.
Conflict of interest statement
Figures


References
-
- Fedullo PF, Auger WR, Kerr KM, Rubin LJ (2001) Chronic thromboembolic pulmonary hypertension. N Engl J Med 345: 1465–1472. - PubMed
-
- Peacock A, Simonneau G, Rubin L (2006) Controversies, uncertainties and future research on the treatment of chronic thromboembolic pulmonary hypertension. Proc Am Thorac Soc 3: 608–614. - PubMed
-
- Lang I (2010) Advances in understanding the pathogenesis of chronic thromboembolic pulmonary hypertension. Br J Haematol 149: 478–483. - PubMed
-
- Pengo V, Lensing AW, Prins MH, Marchiori A, Davidson BL, et al. (2004) Incidence of chronic thromboembolic pulmonary hypertension after pulmonary embolism. N Engl J Med 350: 2257–2264. - PubMed
-
- Dentali F, Donadini M, Gianni M, Bertolini A, Squizzato A, et al. (2009) Incidence of chronic pulmonary hypertension in patients with previous pulmonary embolism. Thromb Res 124: 256–258. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

