The pain interactome: connecting pain-specific protein interactions
- PMID: 24978826
- PMCID: PMC4247380
- DOI: 10.1016/j.pain.2014.06.020
The pain interactome: connecting pain-specific protein interactions
Abstract
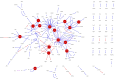
Understanding the molecular mechanisms associated with disease is a central goal of modern medical research. As such, many thousands of experiments have been published that detail individual molecular events that contribute to a disease. Here we use a semi-automated text mining approach to accurately and exhaustively curate the primary literature for chronic pain states. In so doing, we create a comprehensive network of 1,002 contextualized protein-protein interactions (PPIs) specifically associated with pain. The PPIs form a highly interconnected and coherent structure, and the resulting network provides an alternative to those derived from connecting genes associated with pain using interactions that have not been shown to occur in a painful state. We exploit the contextual data associated with our interactions to analyse subnetworks specific to inflammatory and neuropathic pain, and to various anatomical regions. Here, we identify potential targets for further study and several drug-repurposing opportunities. Finally, the network provides a framework for the interpretation of new data within the field of pain.
Keywords: Gene expression; Inflammatory pain; Networks; Neuropathic pain; Protein–protein interactions; Text mining.
Copyright © 2014 The Authors. Published by Elsevier B.V. All rights reserved.
Figures





References
-
- Barabasi A.L., Oltvai Z.N. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. - PubMed
-
- Breivik H., Collett B., Ventafridda V., Cohen R., Gallacher D. Survey of chronic pain in Europe: prevalence, impact on daily life, and treatment. Eur J Pain. 2006;10:287–333. - PubMed
-
- Calvo M., Dawes J.M., Bennett D.L. The role of the immune system in the generation of neuropathic pain. Lancet Neurol. 2012;11:629–642. - PubMed
-
- Cattaneo A. Tanezumab, a recombinant humanized mAb against nerve growth factor for the treatment of acute and chronic pain. Curr Opin Mol Ther. 2010;12:94–106. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

